Figure 4.
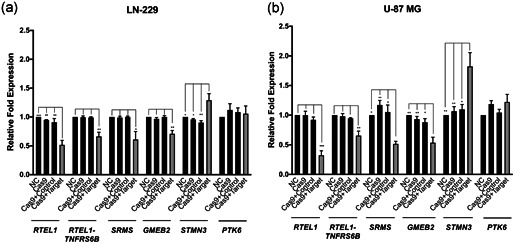
The gene expression changes after CRISPR−Cas9 deletion of Enhancer 2. The genomic region corresponding to enhancer region 2 was targeted for deletion in LN‐229 (a) and U‐87 MG (b) cells using CRISPR−Cas9 technology. Pools of transfected cells were analyzed using qPCR and Taqman gene expression assays for RTEL1, RTEL1‐TNFRSF6B, SRMS, GMEB2, STMN3, PTK6, and TBP(control) custom assays in triplicate, in three independent experiments. cas9, cells transfected with Cas9 only (no guide RNAs); Cas9+Control, cells transfected with Cas9 vector and gRNA empty vector; Cas9+Target, cells transfected with Cas9 vector and guide RNA target vectors; NC, mock‐transfected parental cells. Targeting of the region resulted in a decrease in RTEL1, RTEL1‐TNFRSF6B, SRMS, and GMEB2 expression levels, whereas STMN3 expression levels increased significantly. PTK6 expression levels did not change significantly. *p < .05; **p < .01; and ***p < .001 indicate the levels of significance
