Fig 2. Amino acid differences between orthopoxvirus K3 orthologs.
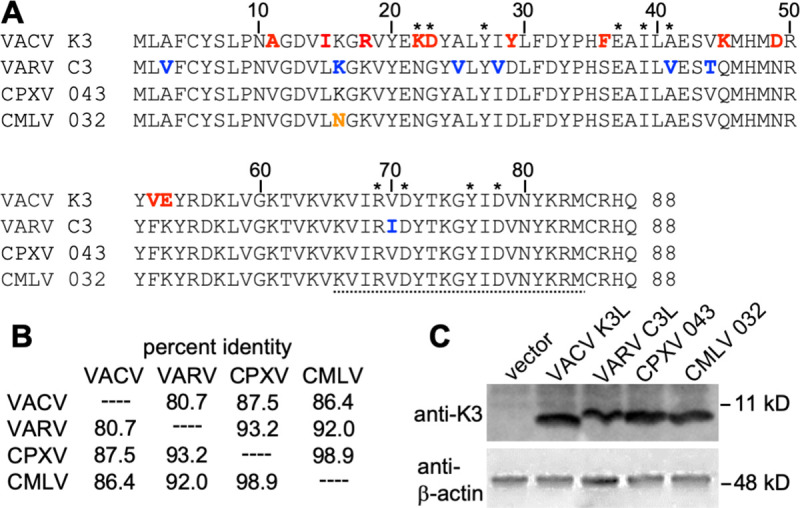
(A) Multiple sequence alignment of VACV K3, VARV C3, CPXV-D 043/TATV 037, and CMLV 032. Amino acids that are unique to VACV K3 are shown in red. Residues that differ between VARV C3 and CMLV 032 are highlighted in blue. The residue that is unique to CMLV 032 is shown in orange. Asterisks denote residues of K3 that correspond to the contacts of eIF2α with PKR in a co-crystal structure [52]. The peptide sequence that was used to generate anti-VACV K3 antibodies is underlined. (B) Amino acid identities between tested K3 orthologs used in this study. Percent identities were calculated from a multiple sequence alignment using MegAlign. (C) Expression of K3 orthologs in transfected cells. HeLa-PKRkd cells were transfected with the indicated K3L orthologs, and cell lysates were collected 48 hours later. Total proteins from these samples were separated on 12% SDS-PAGE gels and analyzed by immunoblot analysis with the indicated anti-K3 and anti-β-actin antibodies.
