Fig. 3. Evidences for oligoamidine 3’s selectivity for bacteria over eukaryotic cells on both of its mechanisms of action.
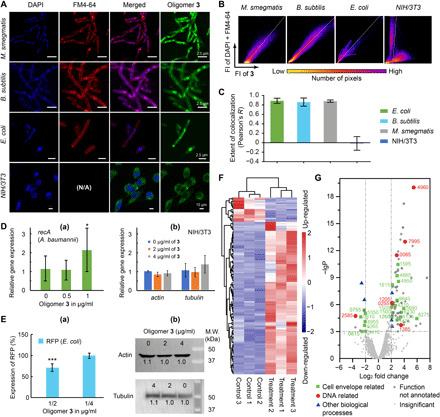
(A) Overlays of confocal microscopic images of bacteria and mammalian cells stained by DAPI, FM4-64, and 3-FITC. N/A, not applicable. (B) Representative two-dimensional fluorescence intensity histograms of stained bacteria and mammalian cells. (C) Pearson’s correlation coefficient, R, for membrane and DNA staining versus staining with 3-FITC. The statistics covered >20 randomly picked cells in each category. (D) Selective activation of housekeeping gene in bacteria cells by 3. (a) 3 activated recA transcription in A. baumannii at its sub-MIC concentration. (b) The transcription of two housekeeping genes in NIH/3T3 cells, actin and tubulin, was not affected at an even higher concentration of 3. (E) Selective inhibition of protein expression in bacteria cells by 3. (a) Oligomer 3 reduced the expression of protein (RFP) in its sub-MIC concentration. (b) Protein expression of Tubulin and Actin in NIH/3T3 cells was not significantly affected. (F) Heatmap of differential expression analysis showing gene regulation changes in A. baumannii treated with 3. Control and treatment groups each had three replicates. (G) Volcano plot of the transcriptome results using 3-treated A. baumannii. -lgP, -log10 (P of the corresponding protein). P is the octanol-water partition coefficient.
