Fig. 2. Construction of cgRNA and scgRNA.
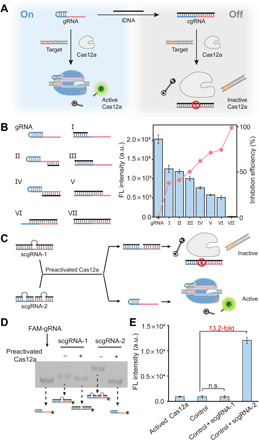
(A) Caging the activity of LbCas12a gRNA using an iDNA. (B) Inhibition on the trans-cleavage activity of LbCas12a by a variety of iDNAs. The 5′-handle and 3′-spacer regions of gRNA are shown in blue and red, respectively. LbCas12a, 50 nM; gRNA, 50 nM; iDNA, 50 nM; target dsDNA (aDNA), 50 nM; FQ reporter, 1.0 μM. FL, fluorescence; a.u., arbitrary units. (C) Design principle of scgRNAs and the release of active gRNA from scgRNA-2. The bulges in scgRNA are shown in gray. (D) Analysis of the cleavage of scgRNA (200 nM) by preactivated LbCas12a (50 nM) using native polyacrylamide gel electrophoresis (PAGE). The gRNA in each scgRNA is labeled by a 6-carboxyfluorescein (FAM-gRNA). (E) Evaluation of the activity of released gRNA from scgRNA-2. Each scgRNA (500 nM) was treated by preactivated LbCas12a (1.0 nM) and then mixed with LbCas12a protein (1.0 μM), aDNA (500 nM), and FQ reporter (1.0 μM). The control sample contains the same components except the scgRNA. Data represent means ± SD. n.s., not significant. Student’s t test.
