Fig. 3. KLF11 and YAP/TEAD bind to joint sites in a cooperative manner.
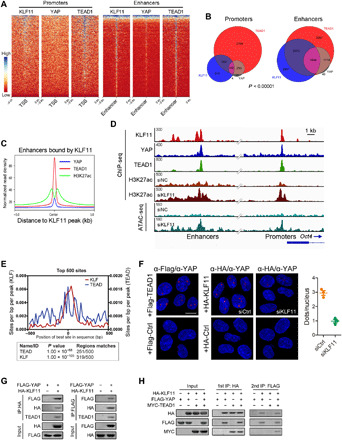
(A) Heatmaps of ChIP-seq data showing the occupancy of KLF11, YAP, and TEAD1 in a ±5-kb window surrounding the center of all RefSeq transcriptional start sites (TSSs) and all enhancer regions in MG63 cells. (B) Venn diagram showing the numbers of promoters and enhancers, respectively, bound by KLF11, YAP and TEAD1in MG63 cells. (C) Density profiles of ChIP-seq data showing the occupancy of YAP, TEAD1, and H3K27ac in a ±5-kb window surrounding the center of KLF11 peaks at enhancer sites. (D) Genome browser view of ChIP-seq and ATAC-seq signals at Oct4 promoters and enhancers as indicated locus in MG63 cells. (E) Centrimo analysis for KLF and TEAD motifs to identify enriched binding motifs surrounding the top 500 KLF11 ChIP-seq peaks. (F) Left: Representative microscopic images from PLA experiments in MG63 sphere-derived cells. PLA signals (red dots) were detected for interaction of endogenous YAP and exogenous FLAG-TEAD (positive control, left) or HA-KLF11 after treatment with siRNAs targeting KLF11 (siKLF11) or control siRNAs (siCtrl). The nuclei were stained with DAPI (blue). Scale bar, 10 μm. Right: Quantification of PLA signals for the YAP1-KLF11 interaction in MG63 sphere-derived cells treated with siKLF11 or siCtrl. (G) Co-IP of exogenous KLF11 and YAP in lysates from human embryonic kidney (HEK) 293T cells overexpressing HA-KLF11 and FLAG-YAP. (H) Two-step co-IP to test for ternary complex formation of KLF11, YAP, and TEAD1. HEK293T cells were transfected with HA-KLF11, FLAG-YAP, and Myc-TEAD1. The first immunoprecipitation was performed with an anti-HA antibody. The complex was eluted with the HA peptide, followed by the second step of immunoprecipitation with an anti-FLAG antibody. The resulting precipitates were analyzed by Western blot analysis with the indicated antibodies. See also figs. S9 and S10.
