Fig. 4. XL-MS analysis of heme-dependent LBD:RID interactions.
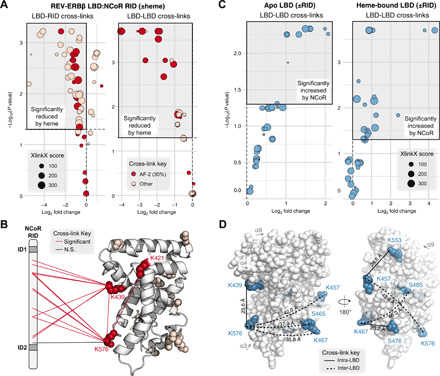
(A) Differential analysis of LBD-LBD and LBD-RID cross-links in REV-ERBβ LBD + NCoR RID samples prepared in the presence or absence of heme. AF-2 cross-links involve residues K421, K439, or K576. The top left quadrant (shaded gray box) highlights cross-links significantly reduced by heme (Padj < 0.05). (B) AF-2 (red) or other (pink) residues involved in cross-links significantly reduced by heme [gray box in (A)] are shown as spheres mapped to PDB 3CQV. LBD-LBD and LBD-RID cross-links involving AF-2 residues are indicated with red or black lines (all but one was significantly reduced by heme). N.S., not significant. (C) Differential analysis of LBD-LBD cross-links in apo (left) or heme-bound (right) REV-ERBβ LBD in the presence or absence of NCoR RID. The top right quadrant (shaded gray box) indicates cross-links significantly increased by RID binding. (D) LBD-LBD cross-links increased by RID in both apo and heme-bound LBD (blue) mapped to PDB 3CQV. Cross-links likely to be inter-LBD (SASD >25 Å) are indicated with dashed lines, while cross-links likely to be intra-LBD (SASD ≤25 Å) are indicated with solid lines; calculated SASDs are shown next to each line.
