A mechanogenetic system was developed using a synthetic gene circuit to produce biologic drugs in response to mechanical loading.
Abstract
Mechanobiologic signals regulate cellular responses under physiologic and pathologic conditions. Using synthetic biology and tissue engineering, we developed a mechanically responsive bioartificial tissue that responds to mechanical loading to produce a preprogrammed therapeutic biologic drug. By deconstructing the signaling networks induced by activation of the mechanically sensitive ion channel transient receptor potential vanilloid 4 (TRPV4), we created synthetic TRPV4-responsive genetic circuits in chondrocytes. We engineered these cells into living tissues that respond to mechanical loading by producing the anti-inflammatory biologic drug interleukin-1 receptor antagonist. Chondrocyte TRPV4 is activated by osmotic loading and not by direct cellular deformation, suggesting that tissue loading is transduced into an osmotic signal that activates TRPV4. Either osmotic or mechanical loading of tissues transduced with TRPV4-responsive circuits protected constructs from inflammatory degradation by interleukin-1α. This synthetic mechanobiology approach was used to develop a mechanogenetic system to enable long-term, autonomously regulated drug delivery driven by physiologically relevant loading.
INTRODUCTION
Smart biomaterials or bioartificial tissues that autonomously respond to biologic cues and drive a therapeutic or restorative response are promising technologies for treating both chronic and acute diseases (1). Mechanotherapeutics, in particular, are a rapidly growing class of smart biomaterials that use mechanical signals or mechanical changes within diseased tissues to elicit a therapeutic response (2–4) and ameliorate the defective cellular mechanical environment (5–8). Current mechanotherapeutic technologies rely on exogenous protein drug delivery or ultrasound stimulation or on synthetic polymer implants that offer a finite life span for drug delivery (9–11). Creating systems with cellular-scale resolution of mechanical forces that offer long-term, feedback-controlled synthesis of biologic drugs could provide a completely new approach for therapeutic delivery.
In contrast to synthetic polymers, biological tissues grow, adapt, and respond to mechanobiologic signals through the use of specialized molecular components, such as mechanically-sensitive ion channels and receptors that transduce specific stimuli from the physical environment (12, 13). In particular, mechanosensitive ion channels are sensitive to both context and deformation mode, making them uniquely suited as mechanotherapeutic sensors (14–16). The transient receptor potential (TRP) family is a class of selective ion channels including some mechanically sensitive members, such as TRPA1, TRP vanilloid 1 (TRPV1), and TRPV4 (17, 18). TRPV4 is activated by osmotic stress and plays an important role in the mechanosensitivity of various tissues such as articular cartilage, uterus, and skin (19–23). In cartilage, TRPV4 has been shown to regulate the anabolic biosynthesis of chondrocytes in response to physiologic mechanical strain (24).
Osteoarthritis is a chronic joint disease for which there are no available disease-modifying drugs, ultimately leading to a total joint replacement once the diseased and degraded cartilage and surrounding joint tissues incapacitate a patient from pain and a loss of joint function (25). Cartilage tissue engineering is a promising strategy to resurface damaged and diseased articular cartilage with an engineered cartilage tissue construct as a means to reduce the need for, or prolong the time before, a total joint replacement (26, 27). An ongoing challenge in the field, however, is developing engineered cartilage constructs that withstand both the high mechanical loads present within the articular joint and the chronic inflammation present within an osteoarthritic joint (28, 29). For example, the knee cartilage of healthy individuals can experience compressive strains of ~5 to 10% during moderate exercise, while individuals with a history of joint injury or high body mass index, populations at risk for developing osteoarthritis, can experience higher magnitudes of cartilage compression under similar loading (30–32). In this regard, a bioartificial tissue that can synthesize biologic drugs in response to inflammation or mechanical loading, either independently or concurrently, could greatly enhance the therapeutic potential of an engineered tissue replacement.
In this study, we engineered a mechanically responsive bioartificial tissue construct for therapeutic drug delivery by using the signaling pathways downstream of the mechano/osmosensitive ion channel TRPV4 to drive synthetic mechanogenetic gene circuits (Fig. 1). To do this, we first establish that mechanical loading of tissue-engineered cartilage activates TRPV4 through fluctuations in the local osmotic environment and not direct mechanical deformation of chondrocytes. We next deconstructed the gene regulatory networks (GRNs) and signaling pathways evoked by TRPV4 activation and revealed the transient activation of several mechanosensitive transcription factors. We engineered synthetic gene circuits to respond to mechanical TRPV4 activation for driving transgene production of an anti-inflammatory molecule, interleukin-1 receptor antagonist (IL-1Ra). While IL-1Ra (drug name anakinra) is approved as a therapy for rheumatoid arthritis and has successfully attenuated osteoarthritis progression in preclinical models, clinical trials of IL-1Ra therapy in patients with established osteoarthritis have not shown efficacy, suggesting that controlled long-term delivery may be necessary for disease modification (33–35). Here, we show that mechanical or osmotic loading of implantable engineered cartilage tissue constructs induces an autonomous mechanogenetic response and protects tissue constructs from inflammatory insult, suggesting a modality for long-term in vivo drug delivery.
Fig. 1. Mechanogenetic transduction and therapeutic drug delivery approach.
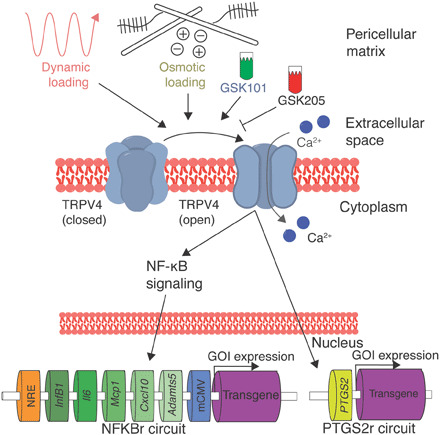
TRPV4 is an osmotically sensitive cation channel in the cell membrane of chondrocytes, which can be activated by mechanical loading secondary to mechano-osmotic coupling through the extracellular matrix or pharmacologically with the agonist GSK101. TRPV4 can also be inhibited with the antagonist GSK205. Upon TRPV4 activation, chondrocytes respond with intracellular calcium signaling that initiates NF-κB signaling and up-regulation of the PTGS2 gene. By lentivirally transducing synthetic mechanogenetic circuits that respond to either NF-κB activation or PTGS2 up-regulation into chondrocytes within an engineered cartilage tissue, mechanically activated TRPV4 signaling was used to drive transgene production of either a luciferase reporter or the therapeutic anti-inflammatory biologic IL-1Ra. GOI; gene of interest.
RESULTS
The mechano-osmotic response of chondrocytes to loading is regulated by TRPV4
To deconstruct the mechanotransduction pathways through which chondrocytes perceive mechanical loading, we encapsulated freshly isolated primary porcine chondrocytes within an agarose hydrogel scaffold to create engineered cartilage tissue constructs. Constructs were cultured for 3 weeks to allow extracellular matrix deposition before applying compressive loading and simultaneously imaging the intracellular calcium levels of the chondrocytes. This engineered cartilage system allows mechanical signals to be transduced to chondrocytes through de novo synthesized extracellular matrix in a manner similar to in vivo mechanotransduction (36–38). Chondrocytes express an array of mechanically sensitive ion channels and receptors, including TRPV4, PIEZO1 and PIEZO2, integrins, and primary cilia (39), so we investigated the mechanism by which physiologic magnitudes of dynamic mechanical loading are transduced by chondrocytes. Dynamic mechanical loading of engineered cartilage constructs immediately provoked a 108% increase of intracellular calcium response compared with unloaded tissues (P = 0.022; Fig. 2A). Inhibiting TRPV4 activity using the TRPV4 antagonist GSK205 suppressed calcium signaling (P = 0.03, GSK205 supplementation reduced signaling by ~47%), implicating TRPV4 as a critical component in transducing compressive loading in chondrocytes.
Fig. 2. Mechanical responsiveness of chondrocytes is mediated by hypo-osmotic stimulation of TRPV4.
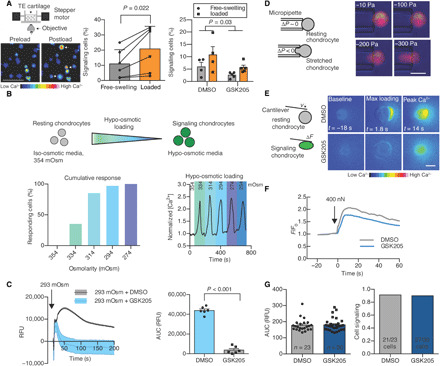
(A) Setup of real-time cellular imaging of mechanical loading. Loading chondrocytes within engineered cartilage increases intracellular calcium compared to free swelling. Arrows indicate immediately responsive cells. Scale bar, 50 μm. The number of cells exhibiting intracellular calcium signaling increased by 108% after loading, and GSK205 suppressed cellular calcium signaling (n = 4 to 5 constructs per treatment). TE, tissue engineered. (B) Isolated chondrocytes are sensitive to osmotic perturbations and exhibit intracellular calcium increases in response to hypo-osmotic stimulation (n = 15 to 20 cells per group, calcium response is normalized to calcium levels at 354 mOsm). (C) Chondrocyte responsiveness to hypo-osmotic stimulation is inhibited with GSK205 (n = 6 per treatment). RFU, relative fluorescence units. (D) Chondrocytes are not sensitive to direct membrane stretch applied under iso-osmotic, iso-volumetric conditions with micropipette aspiration (n = 38; scale bar, 10 μm). (E) Direct cellular compression under a 400-nN load with an AFM induces intracellular calcium signaling. Scale bar, 10 μm. (F) GSK205 does not modulate calcium response of chondrocytes to AFM loading. (G) TRPV4 inhibition alters neither the intensity of calcium responsiveness nor the population response to AFM compression (n = 23 to 30 cells per group). Data are presented as means ± SEM.
While the molecular structure of TRPV4 has recently been reported (40), the mechanisms underlying TRPV4 activation are complex and may involve direct mechanical activation or osmotic activation secondary to mechanical loading of the charged and hydrated extracellular matrix (17, 21, 41, 42). To determine the physical mechanisms responsible for TRPV4 activation secondary to tissue compression, we subjected freshly isolated chondrocytes to osmotic loading, direct membrane stretch, and direct single-cell mechanical compression (Fig. 2, B, D, and E). To better understand the biophysical mechanisms underlying TRPV4 activation, we determined the mechanical state of the chondrocyte membrane in each of these cases using finite element modeling (FEBio; www.febio.org) (43). Chondrocytes rapidly responded to physiologically relevant changes in medium osmolarity through intracellular calcium signals (Fig. 2B). Calcium signaling was highly sensitive to modest changes in osmolarity, with an osmolarity shift of −20 mOsm inducing intracellular signaling in 35% of chondrocytes and −80 mOsm inducing activation of 100% of chondrocytes. Inhibition of TRPV4 with GSK205 suppressed calcium signaling caused by hypo-osmotic loading (Fig. 2C) or by the TRPV4 agonist GSK1016790A (GSK101) (fig. S1). Chondrocyte volumetric analysis and finite element modeling of hypo-osmotic stress showed that a change of −60 mOsm, sufficient to induce signaling in nearly all chondrocytes, increased cellular volume by 13% and induced an apparent first-principle strain of 0.04 homogeneously throughout the membrane (figs. S2 and S3). To investigate the role of direct membrane stretch on TRPV4 activation in the absence of osmotic fluctuations, we used micropipette aspiration of individual chondrocytes to apply controlled deformation of the cell membrane (Fig. 2D). Unexpectedly, micropipette aspiration did not provoke any calcium signaling response in chondrocytes, with only 1 of 38 tested cells responding with increased intracellular calcium, indicating that membrane deformation per se was not the primary signal responsible for mechanical activation. Finite element simulations of the micropipette experiment showed the presence of heterogenous apparent membrane strains in aspirated chondrocytes, with a first-principle strain reaching 0.31 around the micropipette mouth and ~0.04 within the micropipette under an applied pressure of 100 Pa (fig. S3, E to H). To test whether calcium signaling in response to direct mechanical compression of chondrocytes is mediated by TRPV4, we loaded isolated chondrocytes with an atomic force microscope (AFM) to 400 nN (44). Only at high, pathologic levels of mechanical compression did direct chondrocyte compression provoke intracellular calcium signaling, but this response was not inhibited by GSK205 (Fig. 2, E to G). Finite element modeling predicted high membrane strains with an ultimate apparent first-principle membrane strain of 2.27 around the cell at the peak cell deformations necessary to elicit intracellular calcium signaling (fig. S3, I to L). Together, these findings suggest that activation of TRPV4 in chondrocytes in situ may not result directly from cellular strain but rather from osmotic fluctuations induced from the deformation of a chondrocyte’s osmotically active environment. Because of the complex relationship between deformational compressive loading of engineered cartilage and osmotic loading, we will hereafter refer to deformational compressive loading (concomitant with the secondary osmotic effects) as mechanical loading and direct changes in the medium osmolarity as osmotic loading.
TRPV4 activation in chondrocytes induces transient anabolic and inflammatory signaling networks
We next sought to understand the time course of specific downstream signaling pathways and GRNs regulated by mechanical activation of TRPV4 using microarray analysis. Because TRPV4 is a multimodal channel, downstream signaling is likely dependent on the activation mode as well as cell and tissue type. For these studies, primary porcine chondrocytes were cast in agarose to create engineered cartilage constructs and subjected to either compressive mechanical loading [10% sinusoidal peak-to-peak strain at 1 Hz for 3 hours as described in (24, 45)] or pharmacologic stimulation with the TRPV4 agonist GSK101 (1 nM for 3 hours), or left unloaded (free-swelling controls), and measured the transcriptional activity at 0, 3, 12, and 20 hours following an initial round of stimulation and then at 24 and 72 hours after additional daily bouts of mechanical loading or GSK101 (n = 3 per condition per time point; Fig. 3A). In the first 20 hours following initial stimulation, there were 43 transcripts up-regulated under both mechanical loading and pharmacologic stimulation (Fig. 3B and tables S1 and S2) compared to unloaded controls. Up-regulation of these targets was immediate and subsided quickly, with transcript levels returning to baseline 12 to 20 hours after activation. In particular, the adenosine 3′,5′-monophosphate (cAMP)/Ca2+-responsive transcription factors C-JUN, FOS, NR4A2, and EGR2 (Fig. 3C) were all up-regulated in response to TRPV4-mediated calcium signaling.
Fig. 3. Transcriptomic profile induced by TRPV4 activation.
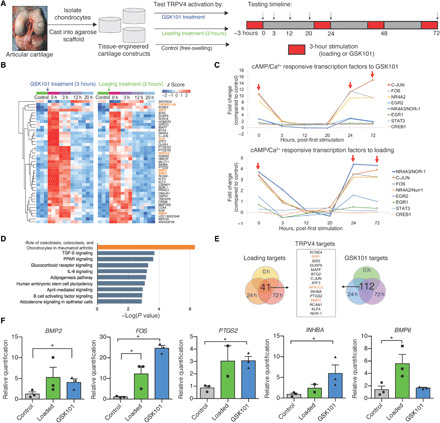
(A) Engineered cartilage tissue constructs were made from isolated primary porcine chondrocytes cast into agarose hydrogels. Tissue constructs were cultured in nutrient-rich medium before deformational mechanical loading or GSK101 pharmacologic stimulation (red, 3 hours per round) following the indicated time course and cartilage construct harvest (arrows). (B) Forty-one genes were differentially up-regulated in response to TRPV4 activation, and levels returned back to baseline after 12 to 20 hours after loading (n = 3 per treatment/time point). (C) cAMP- and calcium-responsive transcription factors were immediately and highly regulated by both mechanical loading and GSK101 stimulation (red arrows indicate removal from loading). (D) Pathway analysis based on transcription activity suggests that both inflammatory and anabolic pathways are strongly regulated by TRPV4 activation. (E) Analysis of gene target response after all bouts of mechanical loading and all bouts of GSK101 stimulation produces a list of distinctly TRPV4-sensitive genes. (F) TRPV4-responsive targets from the microarray analysis were confirmed by qPCR (n = 2 to 3), and a one-tailed t test was used to test whether loaded or GSK101 groups were significantly up-regulated with treatment. *P < 0.05.
We then used Ingenuity Pathway Analysis to identify candidate targets for a synthetic gene circuit that would be responsive to TRPV4 activation. The most enhanced pathway was “the role of osteoblasts, osteoclasts, and chondrocytes in rheumatoid arthritis,” with members including transcription factors (FOS and NFATC2), extracellular matrix synthesis gene (SPP1), growth factors (BMP2 and BMP6), and TNFSF11, the decoy ligand for RANKL, all of which were up-regulated by TRPV4 activation (Fig. 3D). The inflammatory pathways “IL-6 signaling” and “B cell activating factor signaling” were also significantly up-regulated by TRPV4 activation, as were the established chondrocyte anabolic pathways “TGF-β signaling” and “glucocorticoid signaling.” Because of the rapid response times of nearly all of the differentially expressed genes (DEGs), we further analyzed which genes responded to repeated bouts of TRPV4 stimulation at all three time points (0, 24, and 72 hours). There were 41 transcripts repeatedly responsive to mechanical loading and 112 transcripts repeatedly responsive to GSK101 supplementation. Of these targets, 15 genes were commonly responsive to both mechanical loading and pharmacologic activation of TRPV4 (Fig. 3E) and include both pathways associated with inflammation (PTGS2, SPP1, and ATF3) and cartilage development and homeostasis (BMP2, DUSP5, NFATC2, and INHBA). The most robustly regulated and cartilage-relevant genes were confirmed by quantitative polymerase chain reaction (qPCR) (Fig. 3F). Together, these data suggest that the TRPV4 mechanotransduction pathway involves activation of a rapidly resolving inflammatory pathway as part of the broad anabolic response to physiologic mechanical loading.
Synthetic mechanogenetic circuits respond to TRPV4 activation to drive transgene expression
TRPV4 activation in response to mechanical loading up-regulates a diverse group of targeted genes. On the basis of the TRPV4-activated signaling pathway (fig. S4), we identified activation of the nuclear factor κ light chain enhancer of activated B cells (NF-κB) pathway and up-regulation of the prostaglandin-endoperoxide synthase 2 (PTGS2) gene as two distinct avenues to construct TRPV4-responsive synthetic mechanogenetic gene circuits. Targeting the NF-κB signaling pathway and regulation of the PTGS2 promoter (Fig. 1), we developed two lentiviral systems that would either (i) respond to NF-κB activity by linking five synthetic NF-κB binding motifs and a NF-κB–negative regulatory element (46) with the cytomegalovirus (CMV) enhancer to drive transgene expression of either the therapeutic anti-inflammatory biologic IL-1Ra or a luciferase reporter (henceforth referred to as NFKBr-IL1Ra and NFKBr-Luc, respectively) or (ii) respond to PTGS2 regulation by using a synthetic human PTGS2 promoter to drive either IL-1Ra or luciferase expression (henceforth referred to as PTGS2r-IL1Ra and PTGS2r-Luc, respectively). We then created mechanogenetically sensitive engineered cartilage tissue constructs by lentivirally transducing primary porcine chondrocytes with a mechanogenetic circuit and seeding these cells into an agarose hydrogel to produce synthetically programmed cartilage constructs (Fig. 4A).
Fig. 4. Mechanogenetic constructs respond to TRPV4 activation.
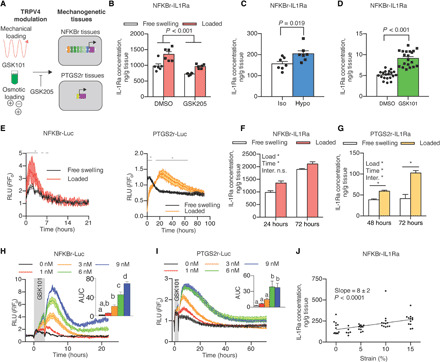
(A) Mechanical loading, osmotic loading, or GSK101 stimulation was applied to mechanogenetic tissues; GSK205 inhibits TRPV4 activation. (B) NFKBr-IL1Ra tissues respond to mechanical loading through increased IL-1Ra (P < 0.001). IL-1Ra is reduced with GSK205 supplementation (P < 0.001, n = 6 per treatment). (C) Exposure of NFKBr-IL1Ra to hypo-osmotic medium produces more IL-1Ra than iso-osmotic medium exposure (P = 0.019, n = 7 per group). (D) NFKBr-IL1Ra tissues exposed to GSK101 stimulation produce more IL-1Ra than vehicle controls (P < 0.001, n = 20 per group). (E) Mechanical loading of NFKBr-Luc tissues quickly activates and inactivates circuits, while PTGS2r-Luc tissues take longer to reach the peak and return to baseline (gray line denotes P < 0.05 between free swelling and load, n = 3 to 6 per group). RLU, relative luminescence units. (F) NFKBr-IL1Ra tissue response to loading after 24 and 72 hours, indicating differential expression in first 24 hours. n.s., not significant. (G) PTGS2r-IL1Ra tissues respond to loading through 72 hours. (H) NFKBr-Luc tissues respond dose dependently to TRPV4 activation via GSK101 through 9 nM GSK101 (P < 0.05, n = 2 to 4 per group). AUC, area under the curve. Different letters denote statistical differences. (I) PTGS2r-Luc tissues are sensitive to GSK101 up to 6 nM (P < 0.05, n = 2 to 4 per group). (J) NFKBr-IL1Ra tissue response is dose dependent to compressive mechanical loading strain from 0 to 15% (P < 0.001, n = 5 to 12). Data are presented as means ± SEM. *P < 0.05.
To test whether mechanogenetic tissues actively respond to mechanical stimulation for transgene production, we first applied compressive mechanical loading to NFKBr-IL1Ra–transduced mechanogenetic cartilage constructs, which showed a 38% increase in IL-1Ra production (P = 0.006) with mechanical loading (Fig. 4B) compared to free-swelling controls. To establish whether this response was dependent on TRPV4, we antagonized TRPV4 with GSK205 and observed a significant attenuation of NFKBr-IL1Ra circuit activity (Fig. 4B), demonstrating that the mechanogenetic circuit responded to both mechanical loading (P < 0.001) and TRPV4 antagonism (P < 0.001). GSK205 also reduced circuit activation in unloaded tissues, suggesting that TRPV4 activation in chondrocytes may not be entirely dependent on mechanical loading. To further assess the specificity of TRPV4 regulation in mechanogenetic cartilage constructs, we applied direct hypo-osmotic loading and pharmacologic GSK101 stimulation to NFKBr-IL1Ra tissues. A step change to hypo-osmotic medium (−200 mOsm change) activated the NFKBr-IL1Ra mechanogenetic circuit (P = 0.019; Fig. 4C) compared to an iso-osmotic medium change (0 mOsm change). In addition, daily pharmacologic activation of TRPV4 with 1 nM GSK101 for 3 hours/day over the course of 5 days activated the NFKBr-IL1Ra circuit compared to dimethyl sulfoxide (DMSO) controls (P < 0.001; Fig. 4D). Engineered cartilage constructs responded repeatedly and reproducibly to the daily GSK101 stimulation (fig. S5), demonstrating the role of TRPV4 in activating the mechanogenetic NFKBr circuits and confirming that a cell-based mechanotherapy can offer prolonged and unabating biologic drug delivery. Testing of conditioned medium from constructs seeded with either nontransduced chondrocytes or chondrocytes transduced with a green fluorescent protein (GFP) expression cassette found that only cells transduced with a mechanogenetic IL-1Ra–producing circuits were capable of synthesizing detectable levels of IL-1Ra.
To determine the sensitivity and temporal response kinetics of our engineered tissue system, factors critical for the effectiveness of any drug delivery system, we used bioluminescence imaging of NFKBr-Luc or PTGS2r-Luc constructs to determine the dynamic response of mechanogenetic cartilage constructs to TRPV4 activation by mechanical loading or GSK101 pharmacologic stimulation. In response to 10% compressive mechanical loading (Fig. 4E), NFKBr-Luc tissue constructs rapidly peaked (1.8 ± 0.2 hours to peak) and decayed (T50% decay time = 3.4 ± 0.4 hours) with loaded samples returning to baseline 4 hours after loading. PTGS2r-Luc tissue constructs were slower to activate (21.7 ± 2.7 hours to peak) and remained activated for a longer duration (T50% decay time = 22.3 ± 1.7 hours). This differential in time delivery kinetics may provide strategies by which mechanical loading inputs can drive both short- and long-term drug production by judicious mechanogenetic circuit selection in a single therapeutic tissue construct. To test whether IL-1Ra production followed similar differential production rates, we mechanically loaded NFKBr-IL1Ra and PTGS2r-IL1Ra tissue constructs and measured protein levels of IL-1Ra released in the medium. One round of mechanical loading activated NFKBr-IL1Ra tissues by 24 hours, as measured by an increase in IL-1Ra produced by loaded tissue constructs compared to unloaded constructs (an increase in IL-1Ra production of 372 ± 265 ng/g, means ± 1 SD). This differential in IL-1Ra concentration between loaded and free-swelling constructs remained unchanged by 72 hours (loaded tissues produced 220 ± 223 ng/g more than unloaded constructs at this time point), indicating that loaded NFKBr tissues were not continually activated and resumed baseline activity levels after 24 hours (Fig. 4F). Conversely, in a preliminary experiment, a single round of mechanical loading did not differentially activate PTGS2r-IL1Ra in the 24 hours after loading (fig. S6). Informed by the bioluminescent imaging, we measured conditioned medium of PTGS2r-IL1Ra tissues 48 and 72 hours after a single round of mechanical loading, however, and found that tissues produced more IL-1Ra after 48 hours compared to free-swelling tissues (difference in means of 21 ng/g) and that the IL-1Ra difference between loaded and free-swelling tissues continued to increase by 72 hours (difference in means of 61 ng/g). This increased effect size confirmed that mechanically loaded PTGS2r tissue constructs remained activated up to 72 hours after loading and demonstrated a longer-acting mechanogenetic response in the PTGS2r system (Fig. 4G). On the basis of the bioluminescent imaging results, we did not measure IL-1Ra levels past 72 hours. These results show that complex drug delivery strategies can be programmed into a single mechanogenetic tissue constructs to produce multiple modes and time scales of therapeutic or regenerative biologic drug delivery.
For insight into the dose response relationship and sensitivity of the mechanogenetic gene circuits, we imaged NFKBr-Luc and PTGS2r-Luc bioluminescence in response to different doses of GSK101. Temporal imaging revealed that GSK101 stimulation produced similar rise and decay kinetics as TRPV4 activation from mechanical loading (Fig. 4E) in NFKBr-Luc (Fig. 4H) and PTGS2r-Luc (Fig. 4I) cartilage tissue constructs compared to unstimulated controls. Both mechanogenetic tissue constructs displayed a clear dose-dependent activation from GSK101 stimulation as demonstrated by the AUC (area under the curve). NFKBr-Luc tissue constructs were responsive from 1 to 9 nM GSK101, whereas PTGS2r-Luc tissue constructs plateaued in response at 6 nM GSK101. On the basis of this pharmacologic sensitivity, we hypothesized that mechanogenetic constructs would be increasingly activated by higher mechanical loading strains through increased osmotic stimulation (Fig. 2B). As NFKBr-Luc tissues demonstrated the most pronounced GSK101 dose-dependent response, we applied dynamic compressive strain amplitudes from 0 to 15% to NFKBr-IL1Ra tissue constructs to span the range of physiologic strains expected for articular cartilage in vivo (36). We measured elevated production of IL-1Ra with increasing compressive strains (slope = 8 ± 2 ng/g IL-1Ra per %strain, P < 0.001; Fig. 4J), demonstrating that drug production is directly responsive to the magnitude of mechanical loading in our mechanogenetic engineered tissues.
Mechanogenetic engineered cartilage activation protects cartilage tissues from IL-1α–induced inflammation-driven degradation
The long-term success of engineered cartilage implants depends on the ability of implants to withstand the extreme loading and inflammatory stresses within an injured or osteoarthritic joint (47). We hypothesized that TRPV4 activation would activate mechanogenetic tissue constructs to produce therapeutic levels of IL-1Ra and protect engineered cartilage constructs and the surrounding joint from destructive inflammatory cytokines. As our mechanogenetic circuits rely on signaling pathways that overlap with the cellular inflammatory response, we examined the dose response of IL-1Ra production in response to the inflammatory cytokine IL-1α (Fig. 5, A and C). NFKBr-IL1Ra mechanogenetic constructs responded to exogenous IL-1α supplementation following a dose-dependent characteristic, consistent with findings of IL-1–induced NF-κB signaling in chondrocytes (Fig. 5B) (48). This dose-dependent response to IL-1α was present up to 10 ng/ml and offered prolonged and robust production of IL-1Ra, promoting the notion that cell-based tissues may offer a more sustained ability to produce therapeutic biomolecules relative to traditional acellular approaches (fig. S7). Mechanical loading further enhanced IL-1Ra production in the presence of IL-1α, demonstrating that, even in the presence of high levels of inflammation, mechanical loading further potentiates NF-κB signaling (fig. S8). Mechanogenetic PTGS2r-IL1Ra and PTGS2r-Luc constructs demonstrated no sensitivity to IL-1α (Fig. 5, D and E). In contrast to NFKBr tissue constructs, the selective sensitivity to TRPV4 activation and not to IL-1α in PTGS2r tissue constructs suggests that this system is distinctively sensitive to mechano- or osmo-activation of TRPV4. This distinction was further demonstrated in tissues exposed to both IL-1α inflammation and osmotic loading, wherein only loading played a significant influence (P = 0.0002 for loading and P = 0.14 for inflammation; fig. S9).
Fig. 5. Activation of TRPV4 via osmotic loading of mechanogenetic constructs protects against IL-1α.
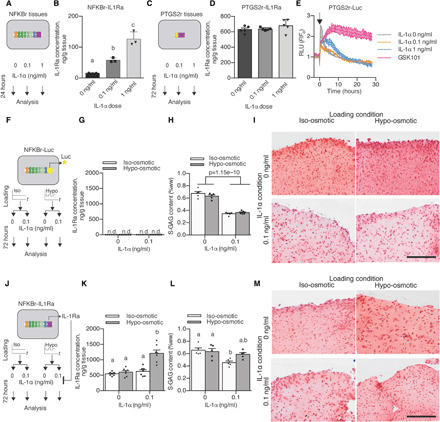
(A) Inflammatory response of NFKBr-IL1Ra constructs under IL-1α supplementation. (B) NFKBr-IL1Ra tissues produce IL-1Ra in response to IL-1α (n = 3, P < 0.001). (C) Inflammatory response of PTGS2r-IL1Ra tissues under IL-1α supplementation. (D) PTGS2r-IL1Ra tissues do not respond to IL-1α (n = 5). (E) PTGS2r-Luc tissues that are not altered by IL-1α are modulated by chronic GSK101 (n = 2 per condition, arrow indicates stimulation). (F) Osmo-inflammatory response of NFKBr-Luc tissues using osmotic loading (3 hours/day) and IL-1α (0 or 0.1 ng/ml) applied to NFKBr-Luc tissues. (G) NFKBr-Luc tissues do not produce IL-1Ra (n = 7). n.d., not detectable. (H) NFKBr-Luc tissues lost S-GAG in the presence of IL-1α (n = 5 per group). (I) Histologically reduced safranin-O staining present with IL-1α supplementation. (J) Osmo-inflammatory response of NFKBr-IL1Ra tissues using osmotic loading (3 hours/day) and IL-1α (0 or 0.1 ng/ml). (K) IL-1Ra was increased with inflammation and osmotic loading (n = 7, P < 0.001). (L) S-GAG content in NFKBr-IL1Ra tissues with IL-1α supplementation (0 or 0.1 ng/ml) and/or osmotic loading (different letters denote significant differences; P < 0.05, n = 5). (M) NFKBr-IL1Ra tissues displayed similar safranin-O staining without IL-1α supplementation, while IL-1α supplementation reduced safranin-O staining in iso-osmotic tissues but not hypo-osmotic tissues. Data are presented as means ± SEM.
To test whether TRPV4 activation–induced IL-1Ra production would protect engineered mechanogenetic tissue constructs against IL-1α, we cocultured mature, 21-day-old NFKBr tissue constructs with articular cartilage explants in the presence of IL-1α inflammation and daily hypo-osmotic loading (incubation in 180 mOsm medium for 3 hours/day; standard iso-osmotic medium was maintained at 380 mOsm) to model the inflammation and osmotic conditions present in the cartilage of arthritic joints exposed to daily loading (Fig. 5, F and J). Using NFKBr-Luc tissue constructs, which lack an anti-inflammatory response to TRPV4 or IL-1α activation (Fig. 5G), to simulate how conventional engineered cartilage constructs would respond in an arthritic joint, constructs lost 45.6% of their sulfated glycosaminoglycan (S-GAG) content after 72 hours of IL-1α treatment (Fig. 5H). S-GAGs are essential structural molecules that impart mechanical integrity and strength in both engineered and native cartilage (49, 50). Osmotic loading did not modulate this response, and the substantial S-GAG loss was observable histologically through diminished safranin-O staining throughout the tissue (Fig. 5I). In the anti-inflammatory NFKBr-IL1Ra tissue constructs, osmotic loading in the presence of inflammatory IL-1α increased IL-1Ra production by 93% over iso-osmotic control tissues also cultured with IL-1α (Fig. 5K). After 72 hours of treatment, IL-1α induced a 30.8% loss of S-GAG in NFKBr-IL1Ra–engineered cartilage constructs under iso-osmotic conditions, while NFKBr-IL1Ra–engineered cartilage that was incubated with IL-1α and osmotically loaded did not significantly lose their S-GAG content (Fig. 5L). Histologically, hypo-osmotic loading NFKBr-IL1Ra–engineered cartilage constructs maintained rich safranin-O staining of S-GAG throughout the constructs, even in the presence of IL-1α (Fig. 5M). Explant proteoglycan levels were similar across all IL-1α treatment groups. Together, these data demonstrate engineered mechanogenetic cartilage constructs that can produce anti-inflammatory IL-1Ra in response to osmotic loading at levels sufficient for engineered tissue protection in a proinflammatory environment mimicking the conditions present in an osteoarthritic joint (51).
DISCUSSION
By combining synthetic biology and tissue engineering, we developed a novel class of bioartificial material that is mechanogenetically sensitive. This system functions by redirecting endogenous mechanically sensitive ion channels to drive synthetic genetic circuits for converting mechanical inputs into programmed expression of a therapeutic transgene. By engineering these cells into a functional tissue construct, this system provides the potential to repair or resurface damaged cartilage while providing site-specific, mechanically induced anti-cytokine therapy against inflammation. Our approach is based on redirection of the downstream response to activation of the TRPV4 ion channel—a critical mechanosensor in cartilage—to transduce tissue-scale deformational mechanical loading via mechano-osmotic coupling in the charged extracellular matrix. By deconstructing the GRNs activated by mechanically induced TRPV4, we coopted chondrocyte-endogenous signaling machinery to drive synthetic circuits for the production of therapeutic biologic drugs, using the anti-inflammatory drug IL-1Ra as a model system for proof of concept. Our results also demonstrate the use of distinct signaling networks for defining the specificity, timing, and dose response for the expression of therapeutic biologic drugs. We show that a single round of mechanical loading can induce short-term and long-term responses based on the particular response of the synthetic circuit. While we targeted a treatment for cartilage repair and osteoarthritis, pathologic mechanical loading and mechano-signaling play a role in a broad range of acute and chronic diseases (12), suggesting a wide range of potential therapeutic applications for mechanogenetically regulated cells or tissues requiring autonomous cellular control systems.
While we developed mechanogenetic cartilage tissues based on TRPV4 activation here, the use of other native, mechanically sensitive ion channels and receptors provides an attractive source of mechanosensors that can be elicited to provoke synthetic outputs. Expanding mechanogenetic approaches to additional mechanosensors with applications to other tissues would increase the range of physical stimuli that synthetic circuits can respond to but requires an in-depth understanding of both the mechanical contexts necessary for mechanosensor activation and the resulting downstream signaling pathways. The fact that primary chondrocytes have an array of different mechanically sensitive ion channels and receptors highlights the potential opportunities to layer mechanosensor-specific circuits and produce systems responsive to different mechanical inputs that drive specific synthetic outputs. Our analysis here demonstrates that physiologic (~10%, 1 Hz) mechanical loading of engineered cartilage is converted to a mechano-osmotic signal that activates TRPV4, evidenced by the GSK205 inhibition of chondrocyte calcium signaling in response to mechanical loading or osmotic loading of engineered cartilage but not to direct cellular deformation or membrane deformation. To this end, we examined the mechanoresponsiveness of TRPV4 signaling and developed two genetic circuits that respond to TRPV4 activation. Note that while endogenous PTGS2 regulation subsided within 24 hours after mechanical loading, our PTGS2r circuit remained activated up to 72 hours after loading, highlighting the role that endogenous mechanisms of gene regulation may play, which are likely absent in our synthetic circuits. In this regard, the use of alternate chondrocyte mechanosensors remains an attractive area of research. For instance, the lack of a TRPV4-dependent response to high-strain compression via AFM loading is consistent with our earlier finding that high, potentially injurious, cellular strains are transduced via the PIEZO family of ion channels (44). The multimodal nature of TRPV4, and the TRP family more generally (52), suggests that additional genetic circuits may be developed for alternate activation modes by characterizing the downstream signaling networks that respond to temperature or biochemical activation of the channel (53). Moreover, engineering novel mechanosensors may open the opportunity for developing custom mechanical activation modes and orthogonal downstream signaling in future mechanogenetic systems.
Our findings offer a detailed perspective of the complex cellular events initiated by TRPV4 activation in chondrocytes. TRPV4 has been thought to play a largely anabolic role in chondrocytes through enhanced synthesis of matrix molecules, S-GAG and collagen, and up-regulation of transforming growth factor–β (TGF-β3) (24), which is evident in our Gene Ontology (GO)/Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway hits of “TGF-β signaling” and “glucocorticoid receptor signaling.” However, our findings also revealed the presence of an acute, transiently resolving inflammatory response (pathways including “chondrocytes in RA,” “IL-6 signaling,” “April-mediated signaling,” and “B cell activating factor signaling”). These findings are particularly striking, as the physiologic levels of mechanical loading applied here (10% compressive strain) are typically associated with promoting chondrocyte anabolism (24), although our data are consistent with other reports suggesting a proinflammatory role of TRPV4 (22, 54, 55). Together, the rapidly resolving mechanical load–induced inflammation in this context may be part of a natural cascade by which a quickly decaying inflammatory response is characteristic of the regenerative, anabolic response to mechanical loading (56, 57). In addition, increasing evidence suggests a potential role for TRPV4 in mediating cellular and tissue inflammation, and studies of chondrocyte-specific TRPV4 knockout mice report decreased severity of age-associated osteoarthritis (54). A load-induced mechanism of chondrocyte inflammation through TRPV4 may provide a target for osteoarthritis and other age-related diseases (22, 55, 58). The synthetic gene circuits developed in this study highlight the opportunities to target different responses through downstream pathway selection, namely, using an NFKBr circuit that is sensitive to IL-1α inflammation and a PTGS2r circuit that is IL-1α insensitive. To this end, the pathologic conditions present in osteoarthritis generate a milieu of inflammatory agents and factors that may, in addition to TRPV4 activation, induce NF-κB signaling and PTGS2 regulation. Deep RNA sequencing and promotor engineering may provide a unique strategy for developing distinct, mechanoresponsive tools in an inflammatory, osteoarthritic joint. Additional therapeutic targets can also be readily inhibited or activated as well; our laboratory has investigated using intracellular inhibitors of NF-κB signaling and the soluble tumor necrosis factor receptor–1 as two alternative options for inhibiting inflammation (59, 60).
It is important to note some potential limitations and future directions of this work. The microarray analysis offers us a first-ever look at the unique transcriptomic landscape induced by TRPV4 activation. To this end, while we used a positive strategy for the groups, i.e., comparing mechanical loading to GSK101 TRPV4 activation, using a negative strategy, i.e., comparing the mechanical loading response to mechanical loading in the presence of GSK205, may offer alternative transcriptomic targets that are TRPV4 regulated. Similarly, with the advent of next-generation sequencing technologies, performing a transcriptomic analysis of the targets of TRPV4 activation would likely increase target resolution, allowing more precise analysis of GRNs induced by TRPV4 activation. Subsequent GRN analysis would permit identification and discrimination between uniquely mechanically responsive and uniquely inflammation-responsive networks to enable our long-term goals toward identifying precise genetic regulatory events driven by TRPV4 activation. Next-generation sequencing technologies may also reveal more robust negatively regulated transcript targets of mechanical loading. Our data here only uncovered several TRPV4-repressed targets, including collagen type VIII α1, suggesting that more refined transcript resolution may be necessary to detect additional targets.
Using an engineered, living tissue construct for coordinated drug delivery obviates many of the traditional limitations of “smart” materials, such as long-term integration, rapid dynamic responses, and extended drug delivery without the need for replacement or reimplantation of the drug delivery system. The modular approach of using cells as both the mechanosensors and the effectors within engineered tissues allows regulation and sensitivity at the mechanically sensitive channel or receptor level, the signal network level, and the gene circuit level. While we used the primary chondrocyte’s endogenous TRPV4 to drive our synthetic system, engineering novel mechanically sensitive proteins may open a new frontier for coordinating inputs or driving receptor activation from novel and precise mechanical inputs. Together, this framework for developing mechanically responsive engineered tissues is a novel approach for establishing new autonomous therapeutics and drug delivery systems for mechanotherapeutics.
MATERIALS AND METHODS
Tissue harvest, cell isolation, agarose gel casting, and culture
Full-thickness porcine articular chondrocytes were enzymatically isolated using collagenase (Sigma-Aldrich) from the femurs of pigs obtained from a local abattoir (~30 kg; 12 to 16 weeks old) post mortem in accordance with an exemption protocol by the Institutional Animal Care and Use Committee. Filtered cells were mixed 1:1 with 4% molten type VII agarose (Sigma-Aldrich), and the cell-agarose mixture was injected into a gel apparatus and allowed to set at room temperature. Chondrocyte-laden disks were punched out, yielding engineered cartilage at a final concentration of 2% agarose and 15 to 20 million cells/ml. All constructs were given 2 to 3 days to equilibrate, and media were changed three times per week during chondrogenic culture using a base medium that consisted of Dulbecco’s modified Eagle’s medium (DMEM) High Glucose (Gibco) supplemented with 10% fetal bovine serum (FBS) (Atlas), 0.1 mM nonessential amino acids (Gibco), 15 mM Hepes (Gibco), proline (40 μg/ml) (Sigma-Aldrich), 1× penicillin/streptomycin, fungizone (Gibco), and fresh ascorbyl-2-phosphate (50 μg/ml) (Sigma-Aldrich) and maintained at 37°C and 5% CO2 (24).
Chondrocyte mechanical and pharmacologic stimulation
Confocal imaging of mechanical compression
A custom mechanical compression device was used to compress agarose constructs while simultaneously performing intracellular calcium imaging on a confocal microscope (Zeiss LSM 880) using fluo-4 AM and fura-red AM (Thermo Fisher Scientific) based on the manufacturer’s protocols. Opposing platens were controlled with stepper motors to apply 60 rounds of compressive loading (10%) after a 2% tare strain. Ratiometric calcium imaging (Calcium Ratio = Intensityfluo-4/Intensityfura-red) was analyzed within each sample with ImageJ for 2.5 min in before and for 2.5 min after the mechanical loading.
High-throughput mechanical compression
A custom mechanical compression device was used for sinusoidal compression of 24 individual tissue constructs simultaneously using a closed-loop displacement feedback system (24). This system allows compression at 37°C and 5% CO2.
Osmotic stimulation
For calcium imaging studies, osmotic loading media were prepared using Hanks’ balanced salt solution medium (Gibco). For mechanogenetic tissue culture studies, osmotic loading media were prepared using DMEM with 1% insulin/transferrin/selenous acid premix (ITS+, Corning, #354352). These media were titrated to hypo-osmotic media by adding distilled water and measured with a freezing point osmometer (Osmette 2007; Precision Systems). For osmotic stimulation of mechanogenetic samples, standard culture medium (containing 10% FBS) was replaced with iso-osmotic (380 mOsm) ITS+ base medium, containing 1% ITS+ premix (Corning), 0.1 mM nonessential amino acids, 15 mM Hepes, and 1× penicillin/streptomycin, 3 days before osmotic loading to acclimate tissues.
Micropipette aspiration
Detailed methods for micropipette aspiration are described previously (61). Briefly, glass micropipettes were drawn to a diameter of ~10 μm and coated with Sigmacote (Sigma-Aldrich) to prevent cell binding to the glass micropipette. The micropipette was brought in contact with a cell, and a tare pressure of 10 Pa was applied for a period of 3 min. Increasing step pressures were then applied in increments of 100 Pa for 3 min each until the cell was fully aspirated. Laser scanning microscopy was used to measure cell deformation [DCI (differential interference contrast)] and [Ca2+]i throughout the experiment. Ratiometric calcium imaging, as described above, was performed to assess the mechanoresponse of chondrocytes to micropipette aspiration; however, we found that the application of step increases in pressure to the cell surface using a micropipette rarely initiated a Ca2+ transient.
Atomic force microscopy
Freshly isolated chondrocytes were plated on glass coverslips and incubated for 2 to 3 days. Before AFM compression, cells were incubated with 10 μM GSK205 or DMSO (vehicle) and stained with intracellular calcium dye Fura-2 AM (Molecular Probes) according to the manufacturer’s protocols. Cells were loaded to 400 nN using an AFM (Asylum Research MFP-3D) with tipless cantilevers (k = 6.2 N/m; MikroMasch) while simultaneously recording intracellular calcium levels with an Olympus microscope and cycled with 340- and 380-nm light to produce a ratiometric output (Calcium Ratio = IntensityFura-2 @ 380 nm/IntensityFura-2 @ 340 nm) for intracellular calcium levels, which were analyzed with ImageJ (44).
Pharmacologic TRPV4 modulation
GSK1016790A (GSK101; MilliporeSigma) was resuspended at final concentrations (1 to 10 nM) and matched with a DMSO (vehicle) control. GSK205 (manufactured at the Duke Small Molecule Synthesis Facility) was used as a TRPV4-specific inhibitor and preincubated with samples before analysis to allow diffusion within three-dimensional tissues and used at a final concentration of 10 μM with appropriate DMSO (vehicle) controls.
FLIPR assay
After digestion and isolation, filtered chondrocytes were plated in a 96-well plate at 10,000 cells per well and left for 24 hours before stimulation on a fluorescent imaging plate reader (FLIPR) by which individual wells were stimulated with either osmotic or GSK101-containing medium. Cellular response in each well was measured via fluo-4 intracellular calcium dye using the Fluo-4 No-Wash Calcium Assay Kit (Molecular Probes) according to the manufacturer’s directions. GSK205 was preincubated and added alongside stimulated and unstimulated chondrocytes to directly assess the TRPV4-dependent response of osmotic and pharmacologic stimulation.
Finite element modeling
Finite element models of cellular deformation were performed using the FEBio (www.febio.org) finite element software package (version 2.6.4). Models were run using a neo-Hookean elastic material for the cell and membrane compartments of the cell (62). All model geometries use axisymmetry boundary conditions to reduce the model size. Osmotic loading was assessed using a Donnan osmotic loading material with parameters taken from the van’t Hoff relation for chondrocytes under osmotic loading and loading chondrocytes with a −60 mOsm osmotic medium shift. Models for micropipette aspiration were run assuming a cell modulus of 1 kPa and Poisson’s ratio of 0.4 while imposing a pressure of −200 Pa to the cell (61). Models of single-cell direct deformational loading (AFM) were performed by simulating an elastic sphere being compressed to 13% of its original height. FEBio testing suites were used to validate the shell, contact, and neo-Hookean code features, and the Donnan model was validated against the van’t Hoff equation.
Microarray collection and analysis
After a 14-day preculture, engineered cartilage constructs (∅4 mm × 2.25 mm) were stimulated under 10% compressive loading or 1 nM GSK101 for 3 hours per day for 3 days. Unstimulated controls (free-swelling) constructs were cultured under identical conditions. Immediately after stimulation, constructs were washed and then fed with culture medium. Constructs were snap-frozen in liquid N2 at 0, 3, 6, 12, 20, 24, and 72 hours after initial stimulation. Total RNA was extracted from the constructs using a pestle homogenizer and the Norgen Biotek RNA/protein purification plus kit. RNA quantity and quality were assessed with NanoDrop (Thermo Fisher Scientific) and the Agilent Bioanalyzer. Total RNA was processed using the Ambion WT expression labeling kit and the Porcine Gene 1.0 ST Array (Affymetrix). The raw signal of arrays was induced into R environment and quantile-normalized by using “affy” and “oligo” package. The significantly DEGs were identified and analyzed by using one regression model in R with package “genefilter,” “limma,” “RUV,” “splines,” “gplots,” and “plotly” at an adjusted P value cutoff of 0.05. Then, the DEGs were imported into Ingenuity Pathway Analysis (IPA) to perform the pathway enrichment analysis. The DEG heatmap was plotted by using gplots in R.
Mechanogenetic circuit design, development, viral development, and culture
We developed two lentiviral systems consisting of an NF-κB–inducible promoter upstream of either IL-1Ra (NFKBr-IL1Ra) or luciferase (NFKBr-Luc). Therefore, upon NF-κB signaling, either IL-1Ra (NFKBr-IL1Ra) or luciferase (NFKBr-Luc) is expressed as a measure of mechanogenetic circuit activation. In addition, we developed two lentiviral systems consisting of a synthetic human PTGS2 promoter upstream of either IL-1Ra (PTGS2r-IL1Ra) or luciferase (PTGS2r-Luc). Therefore, when PTGS2 is activated, either IL-1Ra or luciferase is expressed.
NFKBr circuit design
A synthetic NF-κB–inducible promoter was designed to incorporate multiple NF-κB response elements as previously described (46). A synthetic promoter was developed containing five consensus sequences approximating the NF-κB canonical recognition motif based on genes up-regulated through inflammatory challenge: InfB1, Il6, Mcp1, Adamts5, and Cxcl10 (62). A TATA box derived from the minimal CMV promoter was cloned between the synthetic promoter and downstream target genes, either murine Il1rn or firefly luciferase from the pGL3 basic plasmid (Promega), and an NF-κB–negative regulatory element (NRE-5′-AATTCCTCTGA-3′) was cloned upstream of the promoter to reduce background signal (46, 63, 64).
PTGS2r circuit design
A synthetic human PTGS2 promoter was obtained from SwitchGear Genomics and cloned into the NFKBr-IL1Ra or NFKBr-Luc lentiviral transfer vectors in place of the NF-κB–inducible promoter. The NF-κB–inducible promoter was excised using Eco RI and Psp XI restriction enzymes, and the PTGS2 promoter was inserted in its place using the Gibson Assembly method to create the PTGS2r-IL1Ra and PTGS2r-Luc circuits (65).
Lentivirus production and chondrocyte transduction
Human embryonic kidney (HEK) 293T cells were cotransfected with second-generation packaging plasmid psPAX2 (no. 12260; Addgene), the envelope plasmid pMD2.G (no. 12259; Addgene), and the expression transfer vector by calcium phosphate precipitation to make vesicular stomatitis virus glycoprotein pseudotyped lentivirus (66). The lentivirus was harvested at 24 and 48 hours after transfection and stored at −80°C until use. The expression transfer vectors include the NFKBr-IL1Ra, NFKBr-Luc, PTGS2r-IL1Ra, and PTGS2r-Luc plasmids. The functional titer of the virus was determined with real-time qPCR to determine the number of lentiviral DNA copies integrated into the genome of transduced HeLa cells (66). For chondrocyte transductions, freshly isolated chondrocytes were plated in monolayer at a density of 62,000 cells/cm2 and incubated overnight in standard 10% FBS medium. The following day, virus was thawed on ice and diluted in 10% FBS medium to obtain the desired number of viral particles to achieve a multiplicity of infection of 3 (46). Polybrene was added to a concentration of 4 μg/ml to aid in transduction. The conditioned medium of the chondrocytes was aspirated and replaced with the virus-containing medium, and cells were incubated for an additional 24 hours before aspirating the viral medium and replacing with standard 10% FBS medium. Five days later, cells were trypsinized, counted, and cast in agarose as described above to prepare mechanogenetic constructs at 15 million cells/ml in 2% agarose gel. Constructs were cultured as described above until testing. Viral titers measured by qPCR revealed that ~95% of chondrocytes were transduced with this method.
Mechanogenetic circuit testing outcome measures
Assessing mechanogenetic tissue construct activation in IL-1Ra–producing constructs was measured with an enzyme-linked immunosorbent assay for mouse IL-1Ra (R&D Systems) according to the manufacturer’s protocols. Data are reported as the amount of IL-1Ra produced per construct (in nanograms) normalized by the tissue wet weight mass of the construct (in grams). Luciferase-based mechanogenetic protection was assessed using a bioluminescent imaging reader (Cytation 5, BioTek) at 37°C and 5% CO2 and cultured in phenol red–free high-glucose DMEM supplemented with 1% ITS+, 2 mM GlutaMAX, 1 mM sodium pyruvate, 15 mM Hepes, proline (40 μg/ml), 1× penicillin/streptomycin, and 1 μM luciferin. Before stimulation, samples were imaged under free-swelling conditions for ~1 day to get a baseline bioluminescent level (Fo). For mechanical loading studies, constructs were then mechanically loaded or similarly transferred for a free-swelling control and then returned to the bioluminescent imaging. For GSK101 pharmacologic studies, the bioluminescent medium (above) was supplemented with GSK101 or DMSO (vehicle control) to the appropriate dose (1 to 10 nM) and simultaneously imaged for 3 hours, before washing and replacing with standard bioluminescent medium. Prestimulation baseline was normalized from post-stimulation bioluminescent readings (F) to yield F/Fo as the outcome measure.
Coculture studies
To assess mechanogenetic anti-inflammatory protection, NFKBr-IL1Ra constructs were cultured with a porcine cartilage explant for 72 hours in the presence of porcine IL-1α (0 or 0.1 ng/ml). Porcine cartilage explants (3 mm diameter) were cored from condyle cartilage, and the subchondral bone was removed, leaving a cartilage explant ~1 to 2 mm thick including the superficial, middle, and deep zones and cultured in iso-osmotic ITS+ medium (380 mOsm, formulation listed above) base medium until experimentation. Mechanical loading was applied daily with hypo-osmotic loading (3 hours/day) (180 mOsm) before returning to the iso-osmotic medium (380 mOsm) containing the explant and IL-1α. Iso-osmotic controls were moved similarly to an iso-osmotic medium (380 mOsm). Control, non–anti-inflammatory mechanogenetic tissue constructs consisted of NFKBr-Luc tissues. Tissue S-GAG content in the engineered tissue construct and medium were assessed with the DMB (methylmethylene blue) assay (67). Tissue samples were digested with papain overnight (65°C) to measure tissue S-GAG using the DMB assay. Bulk S-GAG amount was normalized to tissue wet weight. Constructs were fixed in neutral buffered formalin overnight before embedding in paraffin and sectioning to 7 μm. Histological slices were stained with safranin-O to examine S-GAG distribution and abundance.
Statistical analysis
Data were analyzed with one-way analysis of variance (ANOVA) or two-way ANOVA (α = 0.05) where appropriate using R software (www.r-project.org). For two-way ANOVAs, individual groups were compared using a Tukey post hoc analysis when the interaction of factors was also significant. Correlation trends of strain magnitude to IL-1Ra production was analyzed in R using linear model analysis. Temporal bioluminescent data were analyzed using a two-tailed t test comparison between the groups at each analyzed time point.
Acknowledgments
We thank J. Brunger for his contribution in the design of the NFKBr circuits; H. Dressman of the Duke Microarray Facility; I. Rigo and D. Chasse for technical assistance; and G. Erickson, S. Pritchard, and R. Minkhorst for contributions to the cell mechanics assays. Funding: This work was supported by the Shriners Hospitals for Children, the NIH (AR76665, AG46927, AG15768, AR072999, AR74240, AR73752, AR074992, AR073221, and AG28716), the Nancy Taylor Foundation, the Arthritis Foundation, an NSF EAGER Award, the NSF Graduate Research Fellowship Program (DGE-1745038), the Phillip and Sima Needleman Fellowship, the Duke School of Medicine, and Duke Clinical and Translational Science Award (CTSA) UL1TR001117. Author contributions: R.J.N., L.P., and F.G. conceived the project. R.J.N., L.P., N.B.H., A.S., A.K.R., C.J.O., B.Z., A.L.M., and F.G. designed the experiments. R.J.N. and A.S. conducted the chondrocyte mechanical modulation experiments and the finite element modeling. N.B.H., C.J.O., W.B.L., B.Z., and A.L.M. planned, collected, and analyzed the microarray data of TRPV4 stimulation. R.J.N., L.P., J.L., S.S., J.K., and A.K.R. conducted mechanogenetic circuit experiments and analysis. R.J.N., L.P., and F.G. wrote the manuscript. All authors read, edited, and approved the final manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The microarray data generated and analyzed are publicly available (GEO GSE165027). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/5/eabd9858/DC1
REFERENCES AND NOTES
- 1.Lu Y., Aimetti A. A., Langer R., Gu Z., Bioresponsive materials. Nat. Rev. Mater. 2, 16075 (2017). [Google Scholar]
- 2.Intengan H. D., Schiffrin E. L., Structure and mechanical properties of resistance arteries in hypertension: Role of adhesion molecules and extracellular matrix determinants. Hypertension 36, 312–318 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Plodinec M., Loparic M., Monnier C. A., Obermann E. C., Zanetti-Dallenbach R., Oertle P., Hyotyla J. T., Aebi U., Bentires-Alj M., Lim R. Y. H., Schoenenberger C.-A., The nanomechanical signature of breast cancer. Nat. Nanotechnol. 7, 757–765 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Anderson D. D., Chubinskaya S., Guilak F., Martin J. A., Oegema T. R., Olson S. A., Buckwalter J. A., Post-traumatic osteoarthritis: Improved understanding and opportunities for early intervention. J. Orthop. Res. 29, 802–809 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larsen M. B., Boydston A. J., Successive mechanochemical activation and small molecule release in an elastomeric material. J. Am. Chem. Soc. 136, 1276–1279 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Wang J., Kaplan J. A., Colson Y. L., Grinstaff M. W., Mechanoresponsive materials for drug delivery: Harnessing forces for controlled release. Adv. Drug Deliv. Rev. 108, 68–82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu L., Zhang S. X., Liao W., Farhoodi H. P., Wong C. W., Chen C. C., Ségaliny A. I., Chacko J. V., Nguyen L. P., Lu M., Polovin G., Pone E. J., Downing T. L., Lawson D. A., Digman M. A., Zhao W., Mechanoresponsive stem cells to target cancer metastases through biophysical cues. Sci. Transl. Med. 9, eaan2966 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan Y., Yoon S., Sun J., Huang Z., Lee C., Allen M., Wu Y., Chang Y. J., Sadelain M., Shung K. K., Chien S., Wang Y., Mechanogenetics for the remote and noninvasive control of cancer immunotherapy. Proc. Natl. Acad. Sci. U.S.A. 115, 992–997 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohanraj B., Duan G., Peredo A., Kim M., Tu F., Lee D., Dodge G. R., Mauck R. L., Mechanically-activated microcapsules for “on-demand” drug delivery in dynamically loaded musculoskeletal tissues. Adv. Funct. Mater. 29, 1807909 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gossweiler G. R., Hewage G. B., Soriano G., Wang Q., Welshofer G. W., Zhao X., Craig S. L., Mechanochemical activation of covalent bonds in polymers with full and repeatable macroscopic shape recovery. ACS Macro Lett. 3, 216–219 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Di J., Yao S., Ye Y., Cui Z., Yu J., Ghosh T. K., Zhu Y., Gu Z., Stretch-triggered drug delivery from wearable elastomer films containing therapeutic depots. ACS Nano 9, 9407–9415 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Ingber D., Mechanobiology and diseases of mechanotransduction. Ann. Med. 35, 564–577 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Lim C. T., Bershadsky A., Sheetz M. P., Mechanobiology. J. R. Soc. Interface 7, S291–S293 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J., Lewis A. H., Grandl J., Touch, tension, and transduction–the function and regulation of Piezo ion channels. Trends Biochem. Sci. 42, 57–71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.W. Lee, F. Guilak, W. Liedtke, Current Topics in Membranes (Elsevier, 2017), vol. 79, pp. 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ranade S. S., Syeda R., Patapoutian A., Mechanically activated ion channels. Neuron 87, 1162–1179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christensen A. P., Corey D. P., TRP channels in mechanosensation: Direct or indirect activation? Nat. Rev. Neurosci. 8, 510–521 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Moore C., Gupta R., Jordt S. E., Chen Y., Liedtke W. B., Regulation of pain and itch by TRP channels. Neurosci. Bull. 34, 120–142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liedtke W., Choe Y., Martí-Renom M. A., Bell A. M., Denis C. S., AndrejŠali, Hudspeth A. J., Friedman J. M., Heller S., Vanilloid receptor-related osmotically activated channel (VR-OAC), a candidate vertebrate osmoreceptor. Cell 103, 525–535 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liedtke W., Tobin D. M., Bargmann C. I., Friedman J. M., Mammalian TRPV4 (VR-OAC) directs behavioral responses to osmotic and mechanical stimuli in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 100, 14531–14536 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phan M. N., Leddy H. A., Votta B. J., Kumar S., Levy D. S., Lipshutz D. B., Lee S. H., Liedtke W., Guilak F., Functional characterization of TRPV4 as an osmotically sensitive ion channel in porcine articular chondrocytes. Arthritis Rheum. 60, 3028–3037 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ying L., Becard M., Lyell D., Han X., Shortliffe L., Husted C. I., Alvira C. M., Cornfield D. N., The transient receptor potential vanilloid 4 channel modulates uterine tone during pregnancy. Sci. Transl. Med. 7, 319ra204 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Suzuki M., Watanabe Y., Oyama Y., Mizuno A., Kusano E., Hirao A., Ookawara S., Localization of mechanosensitive channel TRPV4 in mouse skin. Neurosci. Lett. 353, 189–192 (2003). [DOI] [PubMed] [Google Scholar]
- 24.O’Conor C. J., Leddy H. A., Benefield H. C., Liedtke W. B., Guilak F., TRPV4-mediated mechanotransduction regulates the metabolic response of chondrocytes to dynamic loading. Proc. Natl. Acad. Sci. U.S.A. 111, 1316–1321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Felson D. T., Lawrence R. C., Hochberg M. C., McAlindon T., Dieppe P. A., Minor M. A., Blair S. N., Berman B. M., Fries J. F., Weinberger M., Lorig K. R., Jacobs J. J., Goldberg V., Osteoarthritis: New insights. Part 2: Treatment approaches. Ann. Intern. Med. 133, 726–737 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Cigan A. D., Durney K. M., Nims R. J., Vunjak-Novakovic G., Hung C. T., Ateshian G. A., Nutrient channels aid the growth of articular surface-sized engineered cartilage constructs. Tissue Eng. Part A 22, 1063–1074 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moutos F. T., Glass K. A., Compton S. A., Ross A. K., Gersbach C. A., Guilak F., Estes B. T., Anatomically shaped tissue-engineered cartilage with tunable and inducible anticytokine delivery for biological joint resurfacing. Proc. Natl. Acad. Sci. U.S.A. 113, E4513–E4522 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ousema P. H., Moutos F. T., Estes B. T., Caplan A. I., Lennon D. P., Guilak F., Weinberg J. B., The inhibition by interleukin 1 of MSC chondrogenesis and the development of biomechanical properties in biomimetic 3D woven PCL scaffolds. Biomaterials 33, 8967–8974 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nims R. J., Cigan A. D., Durney K. M., Jones B. K., O’Neill J. D., Law W.-S. A., Vunjak-Novakovic G., Hung C. T., Ateshian G. A., Constrained cage culture improves engineered cartilage functional properties by enhancing collagen network stability. Tissue Eng. Part A 23, 847–858 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lad N. K., Liu B., Ganapathy P. K., Utturkar G. M., Sutter E. G., Moorman C. T. III, Garrett W. E., Spritzer C. E., DeFrate L. E., Effect of normal gait on in vivo tibiofemoral cartilage strains. J. Biomech. 49, 2870–2876 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins A. T., Kulvaranon M. L., Cutcliffe H. C., Utturkar G. M., Smith W. A. R., Spritzer C. E., Guilak F., DeFrate L. E., Obesity alters the in vivo mechanical response and biochemical properties of cartilage as measured by MRI. Arthritis Res. Ther. 20, 232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutter E. G., Liu B., Utturkar G. M., Widmyer M. R., Spritzer C. E., Cutcliffe H. C., Englander Z. A., Goode A. P., Garrett W. E. Jr., DeFrate L. E., Effects of anterior cruciate ligament deficiency on tibiofemoral cartilage thickness and strains in response to hopping. Am. J. Sports Med. 47, 96–103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelletier J. P., Caron J. P., Evans C., Robbins P. D., Georgescu H. I., Jovanovic D., Fernandes J. C., Martel-Pelletier J., In vivo suppression of early experimental osteoarthritis by interleukin-1 receptor antagonist using gene therapy. Arthritis Rheum. 40, 1012–1019 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Chevalier X., Goupille P., Beaulieu A. D., Burch F. X., Bensen W. G., Conrozier T., Loeuille D., Kivitz A. J., Silver D., Appleton B. E., Intraarticular injection of anakinra in osteoarthritis of the knee: A multicenter, randomized, double-blind, placebo-controlled study. Arthritis Rheum. 61, 344–352 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Schieker M., Conaghan P. G., Mindeholm L., Praestgaard J., Solomon D. H., Scotti C., Gram H., Thuren T., Roubenoff R., Ridker P. M., Effects of interleukin-1β inhibition on incident hip and knee replacement: Exploratory analyses from a randomized, double-blind, placebo-controlled trial. Ann. Intern. Med. 173, 509–515 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez-Adams J., Leddy H. A., McNulty A. L., O’Conor C. J., Guilak F., The mechanobiology of articular cartilage: Bearing the burden of osteoarthritis. Curr. Rheumatol. Rep. 16, 451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mauck R. L., Soltz M. A., Wang C. C. B., Wong D. D., Chao P. H. G., Valhmu W. B., Hung C. T., Ateshian G. A., Functional tissue engineering of articular cartilage through dynamic loading of chondrocyte-seeded agarose gels. J. Biomech. Eng. 122, 252–260 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Lee D. A., Noguchi T., Frean S. P., Lees P., Bader D. L., The influence of mechanical loading on isolated chondrocytes seeded in agarose constructs. Biorheology 37, 149–161 (2000). [PubMed] [Google Scholar]
- 39.Guilak F., Nims R. J., Dicks A., Wu C.-L., Meulenbelt I., Osteoarthritis as a disease of the cartilage pericellular matrix. Matrix Biol. 71, 40–50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng Z., Paknejad N., Maksaev G., Sala-Rabanal M., Nichols C. G., Hite R. K., Yuan P., Cryo-EM and x-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms. Nat. Struct. Mol. Biol. 25, 252–260 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loukin S., Zhou X., Su Z., Saimi Y., Kung C., Wild-type and brachyolmia-causing mutant TRPV4 channels respond directly to stretch force. J. Biol. Chem. 285, 27176–27181 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Servin-Vences M. R., Moroni M., Lewin G. R., Poole K., Direct measurement of TRPV4 and PIEZO1 activity reveals multiple mechanotransduction pathways in chondrocytes. eLife 6, e21074 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maas S. A., Ellis B. J., Ateshian G. A., Weiss J. A., FEBio: Finite elements for biomechanics. J. Biomech. Eng. 134, 011005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee W., Leddy H. A., Chen Y., Lee S. H., Zelenski N. A., McNulty A. L., Wu J., Beicker K. N., Coles J., Zauscher S., Grandl J., Sachs F., Guilak F., Liedtke W. B., Synergy between Piezo1 and Piezo2 channels confers high-strain mechanosensitivity to articular cartilage. Proc. Natl. Acad. Sci. U.S.A. 111, E5114–E5122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng K. W., Mauck R. L., Wang C. C., Kelly T. A., Ho M. M., Chen F. H., Ateshian G. A., Hung C. T., Duty cycle of deformational loading influences the growth of engineered articular cartilage. Cell. Mol. Bioeng. 2, 386–394 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pferdehirt L., Ross A. K., Brunger J. M., Guilak F., A synthetic gene circuit for self-regulating delivery of biologic drugs in engineered tissues. Tissue Eng. Part A 25, 809–820 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng K. W., Lima E. G., Bian L., O’Conor C. J., Jayabalan P. S., Stoker A. M., Kuroki K., Cook C. R., Ateshian G. A., Cook J. L., Hung C. T., Passaged adult chondrocytes can form engineered cartilage with functional mechanical properties: A canine model. Tissue Eng. Part A 16, 1041–1051 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chowdhury T., Salter D., Bader D., Lee D., Signal transduction pathways involving p38 MAPK, JNK, NFκB and AP-1 influences the response of chondrocytes cultured in agarose constructs to IL-1β and dynamic compression. Inflamm. Res. 57, 306–313 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Nims R. J., Durney K. M., Cigan A. D., Dusséaux A., Hung C. T., Ateshian G. A., Continuum theory of fibrous tissue damage mechanics using bond kinetics: Application to cartilage tissue engineering. Interface Focus 6, 20150063 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chahine N. O., Chen F. H., Hung C. T., Ateshian G. A., Direct measurement of osmotic pressure of glycosaminoglycan solutions by membrane osmometry at room temperature. Biophys. J. 89, 1543–1550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McNulty A. L., Rothfusz N. E., Leddy H. A., Guilak F., Synovial fluid concentrations and relative potency of interleukin-1 alpha and beta in cartilage and meniscus degradation. J. Orthop. Res. 31, 1039–1045 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leddy H. A., McNulty A. L., Guilak F., Liedtke W., Unraveling the mechanism by which TRPV4 mutations cause skeletal dysplasias. Rare Dis. 2, e962971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.S. Heller, R. O’Neil, TRP ion channel function in sensory transduction and cellular signaling cascades, in Molecular Mechanisms of TRPV4 Gating, W. Liedtke, S. Heller, Eds. (CRC Press/Taylor & Francis, 2007). [PubMed] [Google Scholar]
- 54.O’Conor C. J., Ramalingam S., Zelenski N. A., Benefield H. C., Rigo I., Little D., Wu C. L., Chen D., Liedtke W., McNulty A. L., Guilak F., Cartilage-specific knockout of the mechanosensory ion channel TRPV4 decreases age-related osteoarthritis. Sci. Rep. 6, 29053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye L., Kleiner S., Wu J., Sah R., Gupta R. K., Banks A. S., Cohen P., Khandekar M. J., Boström P., Mepani R. J., Laznik D., Kamenecka T. M., Song X., Liedtke W., Mootha V. K., Puigserver P., Griffin P. R., Clapham D. E., Spiegelman B. M., TRPV4 is a regulator of adipose oxidative metabolism, inflammation, and energy homeostasis. Cell 151, 96–110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu S., Fadoju D., Rezvani G., De Luca F., Stimulatory effects of insulin-like growth factor-I on growth plate chondrogenesis are mediated by nuclear factor-κB p65. J. Biol. Chem. 283, 34037–34044 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu S., Flint J. K., Rezvani G., De Luca F., Nuclear factor-κB p65 facilitates longitudinal bone growth by inducing growth plate chondrocyte proliferation and differentiation and by preventing apoptosis. J. Biol. Chem. 282, 33698–33706 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Cambré I., Gaublomme D., Burssens A., Jacques P., Schryvers N., de Muynck A., Meuris L., Lambrecht S., Carter S., de Bleser P., Saeys Y., van Hoorebeke L., Kollias G., Mack M., Simoens P., Lories R., Callewaert N., Schett G., Elewaut D., Mechanical strain determines the site-specific localization of inflammation and tissue damage in arthritis. Nat. Commun. 9, 4613 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brunger J. M., Zutshi A., Willard V. P., Gersbach C. A., Guilak F., Genome engineering of stem cells for autonomously regulated, closed-loop delivery of biologic drugs. Stem Cell Reports 8, 1202–1213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guilak F., Pferdehirt L., Ross A. K., Choi Y.-R., Collins K. H., Nims R. J., Katz D. B., Klimak M., Tabbaa S., Pham C. T. N., Designer stem cells: Genome engineering and the next generation of cell-based therapies. J. Orthop. Res. 37, 1287–1293 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trickey W. R., Baaijens F. P., Laursen T. A., Alexopoulos L. G., Guilak F., Determination of the Poisson’s ratio of the cell: Recovery properties of chondrocytes after release from complete micropipette aspiration. J. Biomech. 39, 78–87 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Hou J. C., Maas S. A., Weiss J. A., Ateshian G. A., Finite element formulation of multiphasic shell elements for cell mechanics analyses in FEBio. J. Biomech. Eng. 140, 121009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hou C. H., Huang J., Qian R. L., Identification of α NF-κB site in the negative regulatory element (ε-NRAII) of human ε-globin gene and its binding protein NF-κB p50 in the nuclei of K562 cells. Cell Res. 12, 79–82 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Nourbakhsh M., Hoffmann K., Hauser H., Interferon-beta promoters contain a DNA element that acts as a position-independent silencer on the NF-kappa B site. EMBO J. 12, 451–459 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A. III, Smith H. O., Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Salmon P., Trono D., Production and titration of lentiviral vectors. Curr. Protoc. Hum. Genet. Chapter 12, Unit 12.10 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Farndale R. W., Buttle D. J., Barrett A. J., Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim. Biophys. Acta 883, 173–177 (1986). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/5/eabd9858/DC1


