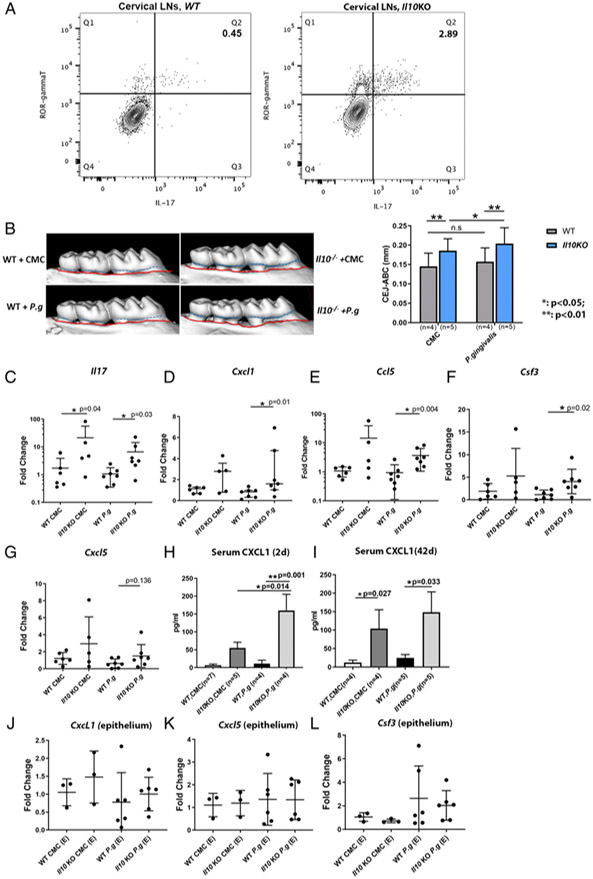
FIGURE 5.
Transcription of IL-17–mediated inflammatory genes in gingival tissues in a P. gingivalis–induced murine alveolar bone-loss model. Flow cytometry plots show the percentages of IL-17–secreting cells (ROR-γt+IL-17+) on previously gated T cells (CD45+CD3+) isolated from WT (A, left panel) and Il10−/− mouse cervical draining lymph nodes (A, right panel) at baseline. Data were from one representative of three separate experiments. Alveolar bone loss from WT and Il10−/− mice that were orally challenged with either sham (CMC) or P. gingivalis is illustrated by the distance between CEJ (outlined by dashed blue line) and ABC (outlined by red line) in representative μCT scans (B, left panel) and compared among different groups (B, right panel). mRNA levels of Il17 (C) and IL-17–associated inflammatory genes, Cxcl1 (D), Ccl5 (E), Csf3 (F), and Cxcl5 (G) in gingival tissues were compared among different groups. Serum levels of CXCL1 from mice 2 d after the last P. gingivalis challenge (H) and 42 d after last challenge (I) were determined by ELISA. mRNA levels of Cxcl1, Cxcl5, and Csf3 from the laser-captured murine gingival epithelia were compared among different groups (J–L). Each experiment was repeated at least twice. Data were pooled for analysis. Each dot represents one sample for mRNA analysis. ANOVA was used to analyze bone loss and serum level of CXCL1, which was shown as the means ± SD. mRNA data were analyzed by Kruskal–Wallis test. *p < 0.05, **p < 0.01.