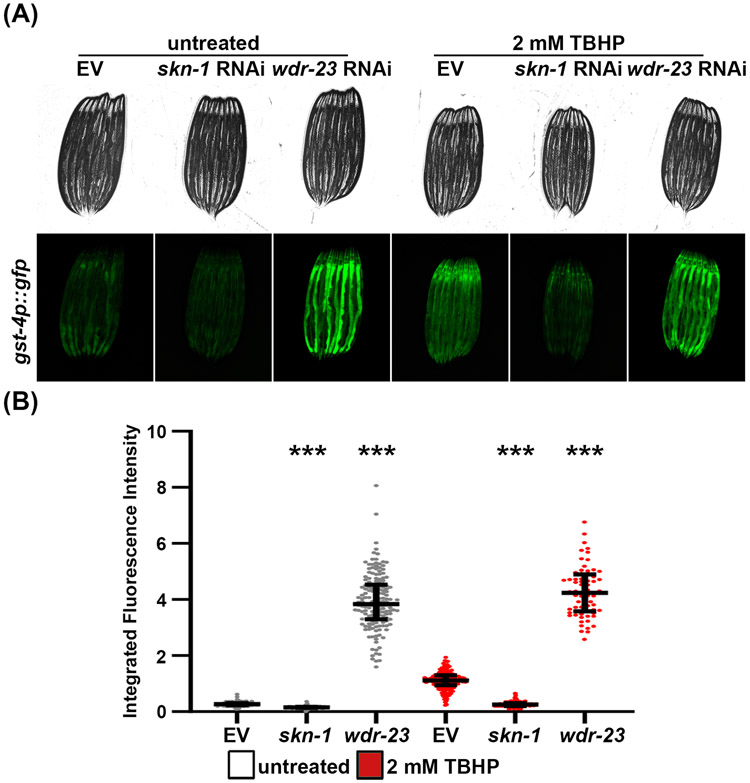
Figure 3. Using gst-4p::GFP as a reporter for the OxSR.
(A) Representative fluorescent micrographs of gst-4p::GFP expressing animals grown on control empty vector (EV), skn-1, or wdr-23 RNAi. Animals were grown on RNAi from hatch until L4 stage at 20 °C. Animals were grown on RNAi from hatch until L4 at 20 °C, then treated with 2 mM TBHP in M9 or only M9 for “untreated” control at 20 °C for 4 hours, and recovered on an EV plate for 16 hours at 20 °C prior to imaging. Animals were paralyzed in 100 μM sodium azide on an NGM agar plate and imaged using a Revolve ECHO R4 compound microscope. (B) Quantitative analysis of (A) using a Union Biometrica large particle biosorter. Data is represented as integrated fluorescence intensity across the entire animal where each dot represents a single animal; untreated control is in grey and TBHP-treated animals are in red. Central line represents the median, and whiskers represent the interquartile range. n = 101-204 animals per strain. *** = p < 0.001 compared to respective EV control using non-parametric Mann-Whitney testing.