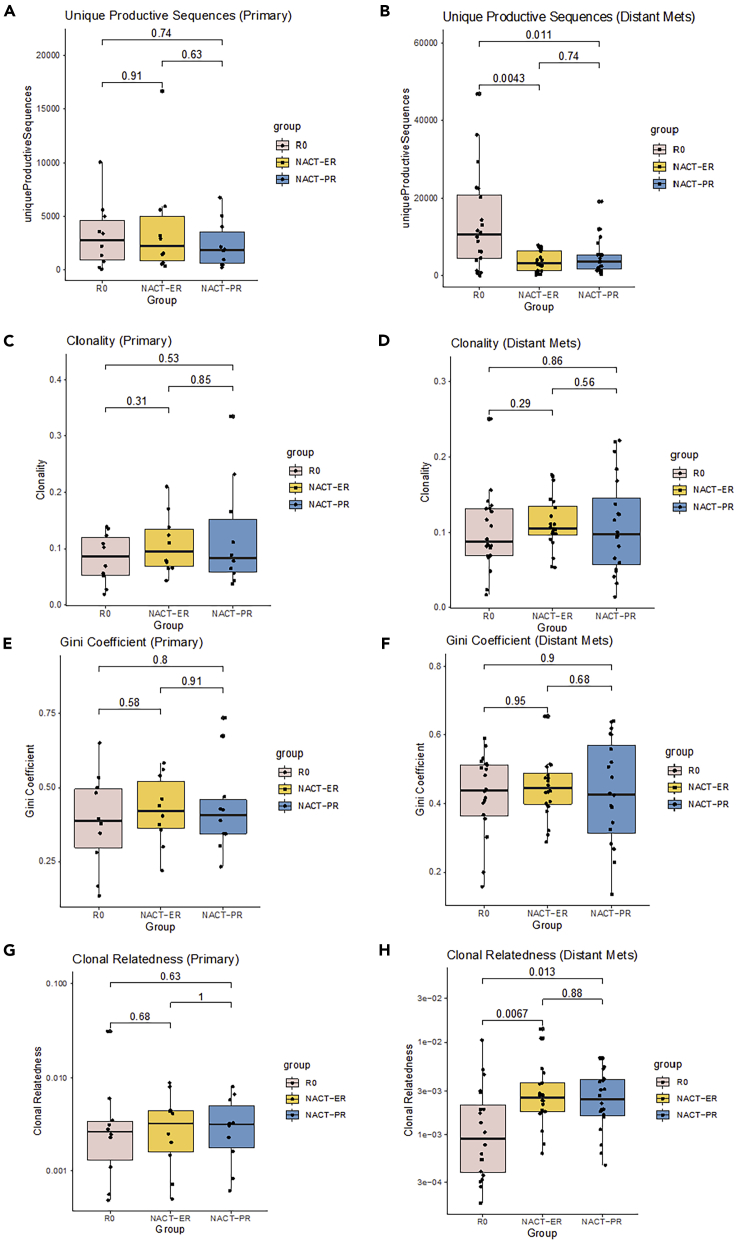
Figure 2.
Diversity evaluation of T cell repertoires by patient group
(A and B) Comparison of T cell repertoire richness in primary tumors (A) and metastatic tumors (B) across three groups. TCR richness was evaluated by the number of unique productive sequences, which were defined as sequences that are in frame and do not have an early stop codon.
(C and D) Comparison of clonality across three groups in primary tumors (C) and metastatic tumors (D). The clonality score is derived from the Shannon entropy, which range from 0 to 1, where 0 indicates that all sequences have the same frequency and 1 indicates that the repertoire is dominated by a single sequence.
(E and F) Comparison of clonal evenness across three groups in primary tumors (E) and metastatic tumors (F). The clonal evenness was evaluated by the Gini coefficient, which range from 0 to 1, where 0 indicates that all clones have identical frequencies, and 1 indicates that they are completely unequal.
(G and H), Comparison of clonal relatedness among three groups in primary tumors (G) and metastatic tumors (H). Clonal relatedness was defined as the proportion of sequences that are related to the most frequent sequence by a defined distance threshold. The value ranges from 0 to 1, where 0 indicates that no sequences are related and 1 indicates that all sequences are related.