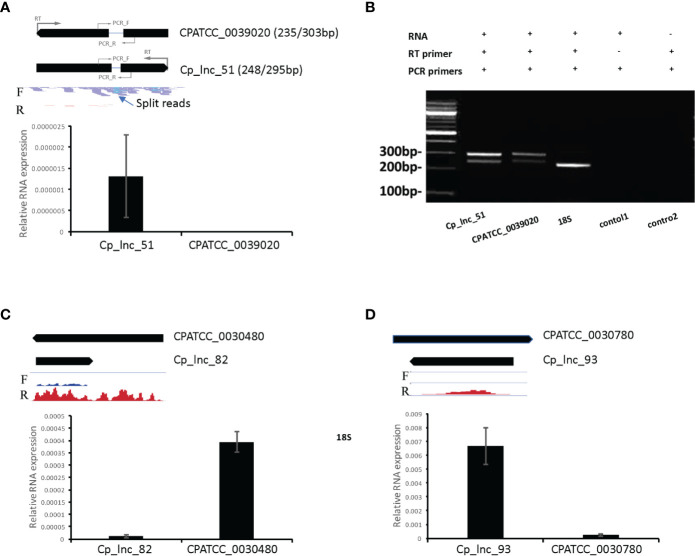
Figure 6.
lncRNA candidate expression and intron structure validation. (A) The expression levels of lncRNA Cp_lnc_51 and its corresponding sense mRNA CPATCC_0039020 were validated by RT-qPCR of RNA from oocyst/sporozoite the expression was normalized to 18S expression levels. The annotated gene model is shown on the top with RNA-Seq reads mapped to the genomic region. Location of RT primers and PCR primers for each gene are shown with the gene models. Reads are separated by the mapped strand: forward strand (F) and reverse strand (R). The intron structure of Cp_lnc_51 is indicated by the presence of split RNA-Seq reads. (B) Agarose gel electrophoresis of RT-PCR products for the sense and antisense transcripts. Splicing of the Cp_lnc_51 transcript and its sense mRNA CPATCC_0039020 are supported by detection of both spliced and unspliced forms for each of the transcripts of the expected sizes. The expected size of the products with/without their intron is indicated next to the gene name in panel A. 18S is used as positive control with expected size of 239bp. Control 1 is a negative control without the RT primer for CPATCC_0039020. Control 2 is a negative control for CPATCC_0039020 with both RT and PCR primers but no RNA template added. (C) The expression levels of lncRNA Cp_lnc_82 and its corresponding sense mRNA CPATCC_0030480 were validated by RT-qPCR. The expression was normalized to 18S expression levels. (D) The expression levels of lncRNA Cp_lnc_93 and its corresponding sense mRNA CPATCC_0030780 validated by RT-qPCR. The expression was normalized to 18S expression levels. The RNA-Seq coverage in C and D is with range 0–100 CPM (counts per million reads mapped).