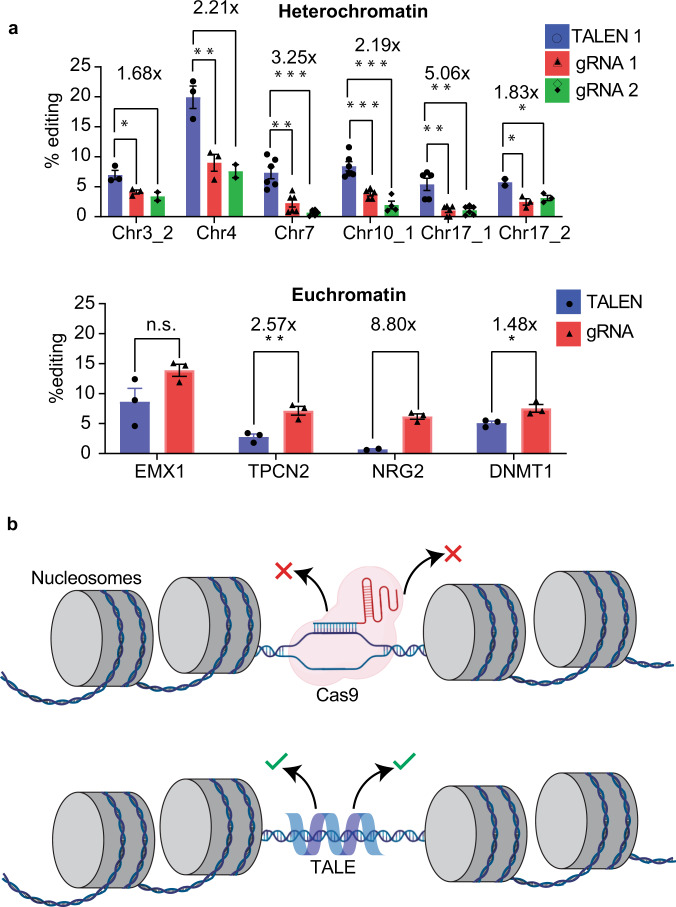
Fig. 5. Systematic comparison of TALE and Cas9 editing efficiency in heterochromatin.
a TIDE analysis of variants of Cas9 and TALEN that target different heterochromatin and euchromatin loci. 50% of the heterochromatin loci are modified more efficiently by TALENs than Cas9 (top). Cas9 has a higher editing efficiency than TALE in the euchromatin regions (bottom). TALENs with the highest predicted cutting efficiency were designed using CHOPCHOP and SAPTA. gRNAs with the highest predicted cutting efficiency were designed using Benchling and CHOPCHOP. TALEN samples are compared to Cas9-gRNA samples by a 2-tailed t test for n > 2. n = {2,3,4,5,6} biological replicates. Data are presented as mean values ± SEM. p < 0.05*, p < 0.01**, p < 0.001***. p-values are given in brackets: Chr3_2 (0.2755, 0.05), Chr4 (0.0096, 0.0171), Chr7 (0.0014, 6.02e-5), Chr10_1 (0.00069, 0.00033), Chr17_1 (0.00179,0.00169), Chr17_2 (0.0027,0.0326), EMX1 (0.0999), TPCN2 (0.0077), NRG2 (0.0025), DNMT1 (0.0274). Source data are provided as a Source Data file. b Illustration of Cas9 and TALE target-search model in heterochromatin. Cas9 becomes trapped in compact heterochromatic structures, whereas TALE can efficiently navigate the heterochromatin.