Abstract
The role of miRNAs in the radiosensitivity of glioma cells and the underlying mechanism is still far from clear. In the present study, we detected six downregulated and seven upregulated miRNAs in the serum after radiotherapy compared with paired serum samples before radiotherapy via miRNA panel PCR. Among these, miR-17-5p was highly reduced (fold change = −4.21). Further, we validated the levels of miR-17-5p in all serum samples with qRT-PCR. In addition, statistical analysis suggested that a reduced miR-17-5P level was positively associated with advanced clinical stage of glioma, incidence of relapse, and tumor differentiation. Moreover, we provided evidence that irradiation markedly activated autophagy and decreased miR-17-5p in the glioma cell line. Further, we demonstrated that irradiation-induced autophagy activation was mediated by beclin-1, and downregulation of beclin-1 via siRNA significantly abolished the irradiation-activated autophagy. Interestingly, we demonstrated that miR-17-5p could directly target beclin-1 via luciferase gene reporter assay. Exotic expression of miRNA-17-5p decreased autophagy activity in vitro. In nude mice, miRNA-17-5p upregulation sensitized the xenograft tumor to irradiation and suppressed irradiation-induced autophagy. Finally, pharmacal inhibition of autophagy markedly enhanced the cytotoxicity of irradiation in RR-U87 cells.
Key words: Glioma, MicroRNA-17-5p, Beclin-1, Autophagy, Radiosensitivity
INTRODUCTION
The incidence of glioma is increasing, and glioma is one of the main causes of cancer-associated deaths in the world (1). This disease recurs in a large number of patients (2). Currently, for postsurgery glioma patients, radiotherapy remains the main complementary method (3,4). However, the efficacy and outcome of radiotherapy are severely limited by radioresistance of glioma cells (5,6). Thus, it is of high necessity to explore the mechanism underlying radioresistance of glioma cells.
MicroRNAs (miRNAs) are highly conserved noncoding small RNAs, which posttranscriptionally regulate the expression of their target genes (7). miRNAs bind to the 3′-untranslated region (3′-UTR) of the target gene mRNA, resulting in destabilization of mRNA and thereby inhibition of protein translation (8). Recently, the role of miRNAs in regulating tumorous radiosensitivity has been intensively studied. miR-21 could modulate the radiosensitivity of cervical cancer through the Akt–mTOR pathway and the PTEN/Akt/HIF-1α loop (9). miR-449a enhances radiosensitivity by downregulation of c-Myc in prostate cancer cells (10). miR-148b is a potential predictor of response to irradiation and could be a promising prognostic biomarker in non-small cell lung cancer (11). However, the potential underlying mechanism of miRNAs in regulating radioresistance of glioma cells remains far from clear.
Autophagy, meaning “self-eating,” refers to a highly conserved catabolic process during which the cells degrade misfolded/dysfunctional proteins and damaged organelles, including the endoplasmic reticulum and mitochondria (12–14). In most circumstances, autophagy acts as an adaptive response to cell stress such as nutrient starvation or metabolic stress (15). Recently, studies show that autophagy is also critical for tumor cellular response to survive stressful conditions and thus has been implicated in the radioresistance of tumor cells (16–19). Thus, targeting autophagy to enhance the radiosensitivity of glioma cells is a promising research direction for improving clinical efficiency and the outcome of radiotherapy.
In the primary study, we detected six downregulated and seven upregulated miRNAs in the serum after radiotherapy compared with paired serum samples before radiotherapy via miRNA panel PCR. Among these, miR-17-5p was highly reduced (fold change = −4.21). Thus, in the present study, we aimed to explore the role of miR-17-5p in the radiosensitivity of glioma cells and the underlying mechanism. Our results indicated that miR-17-5p played an important role in the radiosensitivity of glioma cells by inhibiting beclin-1-mediated autophagy, hinting that the miR-17-5p/beclin-1/autophagy pathway may be a promising therapeutic target to enhance the efficacy of radiotherapy in glioma.
MATERIALS AND METHODS
Cell Culture and Animal Preparation
The U87 cell line, a gift from Prof. Yifan Zhang from the Chinese Academy of Sciences, was cultured in the DMEM (Thermo Fisher Scientific, Cambridge, MA, USA) supplemented with 10% fetal bovine serum (Gibco, Thermo Fisher Scientific) and penicillin/streptomycin. The cells were maintained at 37°C in a humidified incubator with 5% CO2.
Eight-week-old BALB/c female nude mice were purchased from the Experimental Animal Center of The First Hospital of Jilin University (Changchun, P.R. China). All the mice were housed under pathogen-free circumstance and had free access to food and water. The protocol for animal experiments was reviewed and approved by the Animal Care and Use Committee of The First Hospital of Jilin University.
Glioma Specimen Collection
One hundred and ten glioma samples (WHO I–IV) were gathered between January 1, 2006 and January 1, 2014 in The First Hospital of Jilin University, P.R. China. All the samples were classified to WHO I–IV, abiding by the criteria of World Health Organization (WHO) classification. Normal brain tissues were collected in surgery for traumatic brain injury. qRT-PCR was used to determine the miR-17-5p levels between glioma tissue and paired normal brain tissue. If the level of miR-17-5p in the glioma tissue was higher than that in the paired normal brain tissue, we defined this tumor sample as “high” level of miR-17-5p. Otherwise, the tumor was defined as “low” level. The relationship between miR-17-5p level and clinicopathological parameters in glioma patients is shown in Table 2. All patients’ written informed consents were obtained, and the procedures of the present study were reviewed and approved by the Ethical Review Board of The First Hospital of Jilin University, P.R. China.
Table 2.
Association Between miR-17-5p Expression and Clinicopathological Parameters in 110 Patients With Glioma
| Parameters | Total (n) | miR-17-5p Expression | p Value | |
|---|---|---|---|---|
| High [n (%)] | Low [n (%)] | |||
| Total | 110 | 27 (24.54545455) | 83 (75.45454545) | |
| Age | 0.75303 (NS) | |||
| <60 | 42 | 11 (26.19047619) | 31 (73.80952381) | |
| >60 | 68 | 16 (23.52941176) | 52 (76.47058824) | |
| Gender | 0.83018 (NS) | |||
| Male | 51 | 13 (25.49019608) | 38 (74.50980392) | |
| Female | 59 | 14 (23.72881356) | 45 (76.27118644) | |
| Clinical stage | 0.00189 | |||
| I–II | 57 | 21 (36.84210526) | 36 (63.15789474) | |
| III–IV | 53 | 6 (11.32075472) | 47 (88.67924528) | |
| Relapse | 0.01089 | |||
| No | 54 | 19 (35.18518519) | 35 (64.81481481) | |
| Yes | 56 | 8 (14.28571429) | 48 (85.71428571) | |
| Tumor differentiation | 0.00028 | |||
| Well | 41 | 18 (43.90243902) | 23 (56.09756098) | |
| Moderate | 69 | 9 (13.04347826) | 60 (86.95652174) | |
NS, not significant.
Generation of Radioresistant Cell Line
U87 cells in the phase of exponential proliferation were exposed to irradiation at a dose of 6.4 Gy with a high-energy X-ray accelerator (2 Gy/min). In order to generate the radioresistant cell line, the cells were irradiated for eight rounds. After treatment, the remaining cells were designated as radioresistant U87 (RR-U87) cells.
Serum miRNA Microarray
We used miRCURY LNA™ microRNA Arrays (Exiqon, Shanghai, P.R. China) to identify the global miRNA profiling from the serum. RNAs were purified from paired serum samples before and after radiotherapy (BR and AR) from 53 patients. The age range of these patients was 54 to 66 years. All samples were qualified for the quality control analysis and showed no PCR inhibition. The microarrays contained ∼1,200 assay probes corresponding to the annotated human and nonhuman primate miRNA sequences used to determine the differentially expressed miRNAs in the present study. The RNAs were labeled and hybridized according to the manufacturer’s instructions.
qRT-PCR for miR-17-5p
qRT-PCR using TaqMan MicroRNA assays (Applied Biosystems, Shanghai, P.R. China) was performed to evaluate the miR-17-5p and their precursor levels in this study. The qRT-PCR was performed with an ABI 7500 instrument (Applied Biosystems). U6 or miR-16 was used as internal control in the cell samples or serum samples, respectively, and measured by the same method. Briefly, 5 ng of small RNA or total RNA was reverse transcribed and was then amplified with specific primers and TaqMan probes through PCR. The thermocycler conditions used were as follows: step 1, 95°C for 2 min; step 2, 35 cycles at 95°C for 15 s and 55°C for 1 min. The results from qRT-PCR were analyzed using the equation 2−ΔΔCt. Primers used are as follows: miR-17-5p, 5′-GAATCGACATAGCCAAAAAAGTACGACA-3′ (forward) and 5′-ACTATACGTAACACGTAAACATTCGCC-3′ (reverse); miR-16, 5′-GTATAGCTAGCTAGGTAGAGGA-3′ (forward) and 5′-ACGTCCATGCAGCCCTCATGTCTA-3′ (reverse); control U6: 5′-CTCGCT TCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATT TGCGT-3′ (reverse).
Western Blot Analysis
The protein samples were prepared with RIPA lysis buffer supplemented with protease inhibitor and phosphatase inhibitor mix and boiled at 95°C for 10 min. The proteins were separated by 10% SDS gel and transferred to polyvinylidene fluoride (PVDF) membranes in a cold room. After that, the PVDF membranes were blocked with 5% skimmed milk [dissolved in Tris-buffered saline-Tween (TBST)] at room temperature (37°C) for 2 h and then incubated overnight with a primary antibody with gentle shaking at 4°C. After five washes with TBST, the membrane was incubated with a peroxidase-conjugated secondary antibody at room temperature (37°C) for 2 h. Following five washes with TBST, the target protein was visualized with chemiluminescence (ECL) (Thermo Fisher Scientific, Beijing, China). The primary antibodies used in this study include anti-LC3-I/II, anti-p62, anti-beclin-1, and anti-Atg7 (Sigma-Aldrich, St. Louis, MO, USA) and anti-cleaved caspase 3 (Abcam, Cambridge, UK). GAPDH (Abcam) was used as an internal control.
MTT Assay
U87 cells (5 × 103), which are pretreated with 4 μM 3-methyladenine (3-MA) or 20 μM chloroquine (CQ) for 1 h (Beyotime Biotechnology, Wuhan, China) and then exposed to irradiation (2 Gy/min), were seeded in a 96-well plate. At 24 and 48 h posttreatment, the viability of cells was tested by MTT assay as previously described (20). Briefly, 10 μl of 5 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) labeling reagent was added into each well (100 μl of medium) of a 96-well plate, and the U87 cells in the 96-well plate were further cultured at 37°C in a humidified incubator for 4 h. After gentle shaking for 30 s, the absorbance of each well was determined at a wavelength of 570 nm using SPECTROstar Nano spectrometer (BMG LABTECH, German).
TUNEL Assay
Apoptosis was determined with a TUNEL kit (Thermo Fisher Scientific, Shanghai, China) according to the manufacturer’s instructions. Briefly, U87 cells were cultured on coverslips and treated with 4 μM 3-MA or 20 μM CQ for 1 h. At 48 h posttreatment, the cells were fixed with 4% paraformaldehyde (PFA) solution for 10 min at room temperature (37°C). Then in order to block endogenous peroxidase activity, the cells were exposed to 500 μl of methanol with 0.3% H2O2 for 20 min at 37°C. After that, the cells were incubated with TUNEL reaction mixture at 37°C for 60 min and visualized with confocal microscopy (A1; Nikon, Japan).
Transfection With Small Interfering RNA
Cells were seeded in 100-mm plates. When the confluence of the cells reached 70%, the transfection of 100 nM small interfering RNA or scramble siRNA was performed with Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. At 6 h posttransfection, the opti-MEM was replaced with normal medium, and cells were cultured for an additional 24 h. The siRNAs bought from GenePharma (Shanghai, China) are as follows: si-beclin-1 (5′-CAGTTTGGCACAATCAATA-3′) and si-control (5′-GAGAGCUAGACAAUGAAG-3′).
Dual-Luciferase Reporter Assay
The 3′-UTR of beclin-1 and the mutated 3′-UTR constructs were amplified and inserted into pGL3 Luciferase Promote Vector (Sangon, Wuhan, China) with XbaI and NotI restriction sites. The pGL3 vector containing wild-type or mutated 3′-UTR of beclin-1 was cotransfected with miR-17-5p mimics (Sangon) into U87 cells with Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instruction. At 48 h after transfection, the luciferase activity of the cells was determined with the Pierce Renilla-Firefly Luciferase Dual Assay Kit (Thermo Fisher Scientific).
miR-17-5p Mimics
mirVana miRNA mimics (50 nM) for miR-17-5p or scramble molecules (50 nM) were purchased from Sangon and transfected into U87 cells with Lipofectamine 2000 (Thermo Fisher Scientific).
Tumorigenesis
U87 cells (5 × 107) infected with lentiviral vector expressing miRNA-17-5p or scramble miRNA control (Sangon) were injected subcutaneously into the left groin of 8-week-old BALB/c female nude mice. When the tumors reached about 500 mm3 on day 15, a single dose of 10 Gy irradiation was given. The volume of the tumor was measured every day, and the tumor was carefully dissected at 40 days following inoculation. The volume was calculated using the formula: length × width2 × π/6.
Statistical Analysis
All the data were presented as mean ± SD. Chi-square and Fisher’s exact tests were used to analyze the difference between categorical variables. Two-tailed Student’s t-test or one-way ANOVA was used to analyze the difference between or among groups, respectively. All the statistical analysis was performed with SPSS 19.0 software (SPSS Inc., Chicago, IL, USA). A value of p < 0.05 was considered to be statistically significant.
RESULTS
miRNAs Expression in Paired Serum Samples (Before and After Radiotherapy) and the Relationship With Clinical Features
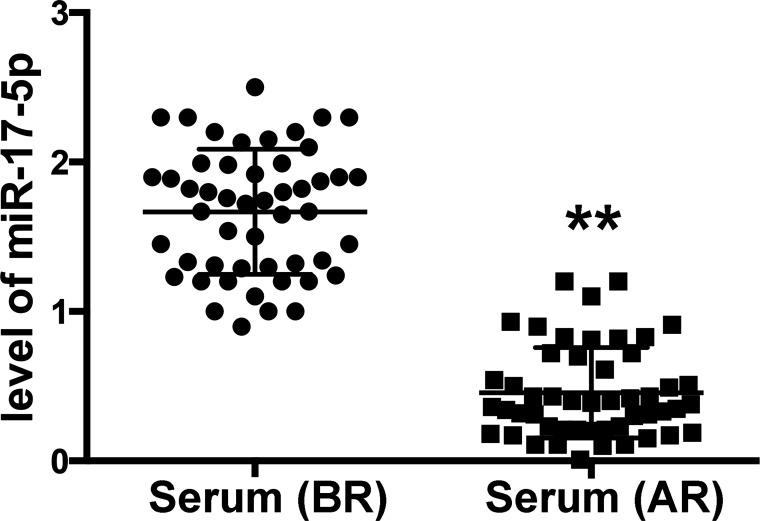
In our primary study, compared with paired serum samples before radiotherapy, six downregulated and seven upregulated miRNAs were identified in the serum after radiotherapy by miRNA panel PCR (fold change >1.5) (Table 1). Among these, miR-17-5p was highly downregulated after irradiation (fold change = −4.21). Further, we validated the levels of miR-17-5p in all serum samples with qRT-PCR. As expected, miR-17-5p was significantly decreased in serum after radiotherapy compared to paired serum before radiotherapy (Fig. 1). Finally, we explored the association between miR-17-5p levels and clinical features. As shown in Table 2, statistical analysis suggested that reduced miR-17-5P level was positively associated with the advanced clinical stage of glioma, incidence of relapse, and tumor differentiation.
Table 1.
miRNAs Differentially Expressed in Paired Serum Samples (Before and After Radiotherapy)
| No. | miRNA Name | Mean Fold Change | p Value |
|---|---|---|---|
| 1 | miR-17-5p | −4.21 | 0.0063 |
| 2 | miR-125 | −3.67 | 0.0123 |
| 3 | miR-126 | −2.97 | 0.0041 |
| 4 | miR-200c | −2.34 | 0.0387 |
| 5 | miR-787 | −1.87 | 0.0481 |
| 6 | miR-24 | −1.65 | 0.0091 |
| 7 | miR-134 | 3.41 | 0.0002 |
| 8 | miR-155 | 3.01 | 0.0013 |
| 9 | miR-335 | 2.56 | 0.0312 |
| 10 | miR-503 | 2.31 | 0.0198 |
| 11 | miR-328 | 1.97 | 0.0374 |
| 12 | miR-101 | 1.89 | 0.0422 |
| 13 | miR-483-3p | 1.68 | 0.0175 |
Positive and negative fold change scores mean significant upregulation and downregulation, respectively, in serum after radiotherapy, compared with that in serum before radiotherapy.
Figure 1.
The level of miR-17-5p in paired serum samples (before and after radiotherapy) and in radioresistant U87 cells. Total RNAs were extracted from the paired serum samples before radiotherapy (BR) and after radiotherapy (AR) from 53 patients with an age range of 54 to 66 years for miRNA panel PCR. After that, the level of miR-17-5p in the serum samples was confirmed with qRT-PCR. Data are presented as mean ± SD. **p < 0.01 compared with the serum before radiotherapy.
Irradiation Activated Autophagy and Reduced miR-17-5p in U87 Cells
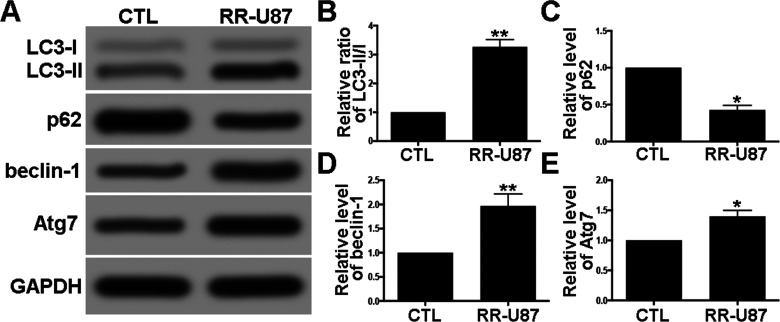
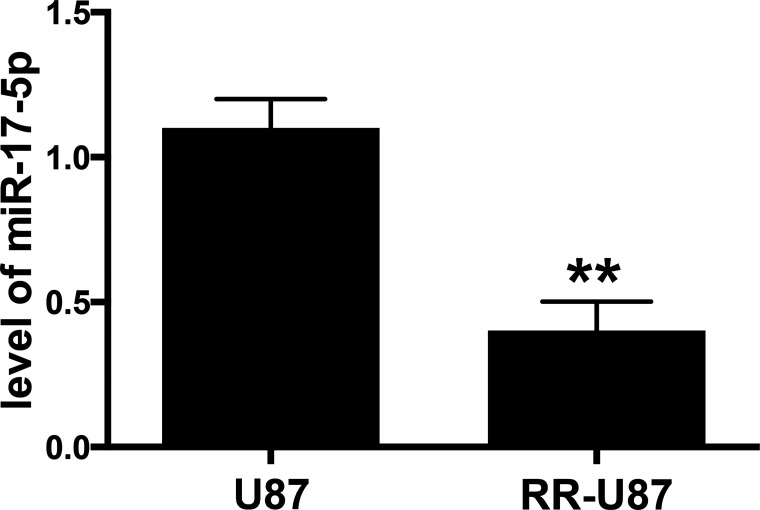
In order to explore the potential role of miR-17-5p in radioresistance of glioma cells, we established a radioresistant U87 (RR-U87) cell line. Interestingly, a significant increase in ratio of LC3-II/I, beclin-1, and Atg7, as well as a decrease in selective autophagy target p62 were detected in RR-U87 cells (Fig. 2A–E). We further examined the level of miR-17-5p in radioresistant cells. Compared with U87 cells without irradiation, miR-17-5p was markedly reduced in RR-U87 cells (Fig. 3). The above data demonstrated that irradiation significantly reduced miR-17-5p and activated autophagy in U87 cells.
Figure 2.
Irradiation-activated autophagy in U87 cells. (A) Autophagy activity was detected by Western blot for LC3-I/II, p62, beclin-1, and Atg7. (B–E) Relative levels of ratios of LC3-II/I (B), p62 (C), beclin-1 (D), and Atg7 (E) were measured using ImageJ and normalized to GAPDH. CTL (control) denotes U87 cells without irradiation, and RR-U87 denotes radioresistant U87 cells. Data are presented as mean ± SD. *p < 0.05 and **p < 0.01 compared with control group.
Figure 3.
The level of miR-17-5p in radioresistant U87 cells. Radioresistant U87 (RR-U87) cell line was generated, and the level of miR-17-5p in RR-U87 cells was also determined by qRT-PCR. Data are presented as mean ± SD. **p < 0.01 compared with U87 cells without irradiation.
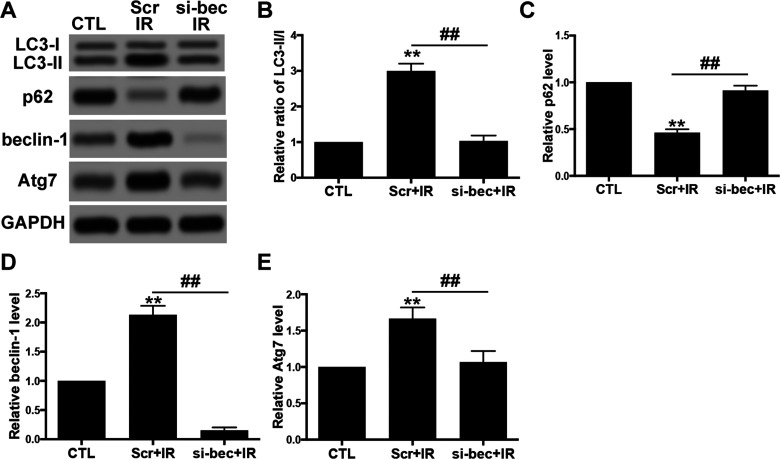
Irradiation-Induced Autophagy Activation Was Mediated by Beclin-1
In order to determine whether irradiation-enhanced autophagy activity was mediated by beclin-1, we cotransfected RR-U87 cells with si-beclin-1 vectors and determined autophagy activity by Western blot. The data revealed that, compared with the scramble group, downregulation of beclin-1 significantly decreased the ratio of LC3-II/I and Atg7 and increased p62 in RR-U87 cells in response to irradiation (Fig. 4A–E). Taken together, the data demonstrated that beclin-1 was the vital mediator for irradiation-induced autophagy.
Figure 4.
Irradiation-induced autophagy activation was mediated by beclin-1. (A) U87 cells were transfected with siRNA for beclin-1 (si-bec) for 24 h and then exposed to irradiation (2 Gy/min). Levels of LC-3I/II, p62, beclin-1, and Atg7 were detected with Western blot. (B–E) Relative levels of ratios of LC3-II/I (B), p62 (C), beclin-1 (D), and Atg7 (E) were measured using ImageJ and normalized to GAPDH. CTL (control) denotes U87 cells without irradiation. Scr denotes scramble. Si-bec denotes siRNA for beclin-1. IR denotes irradiation. Data are presented as mean ± SD from at least three independent experiments. **p < 0.01 compared with control group and ##p < 0.01 compared with the indicated groups.
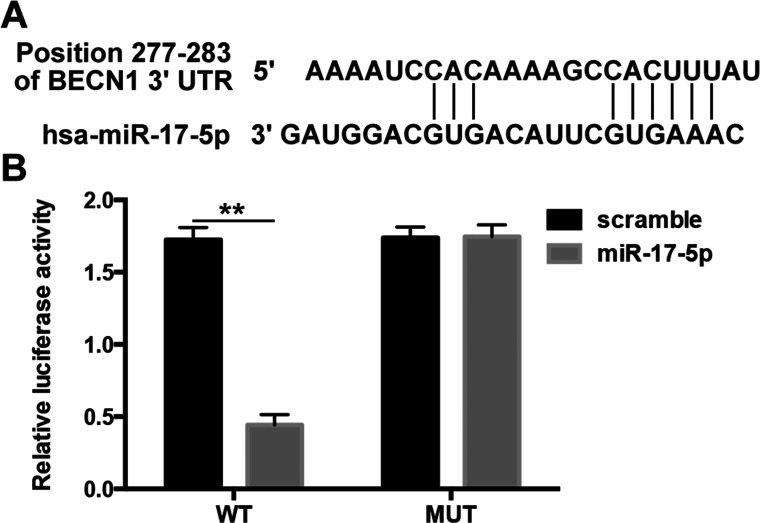
Beclin-1 Is a Direct Target of miR-17-5p
We used TargetScan on the internet to predict the relationship between miRNA-17-5p and beclin-1. As expected, TargetScan showed that miR-17-5p could directly target the 3′-UTR of beclin-1. The binding site between miR-17-5p and beclin-1 3′-UTR is illustrated (Fig. 5A). To validate the physical interaction between miR-17-5p and beclin-1, 3′-UTR of beclin-1 and the mutated 3′-UTR constructs were amplified and inserted into pGL3 Luciferase Promote Vector and cotransfected with miR-17-5p mimics in U87 cells. As expected, miR-17-5p led to a significant reduction in the luciferase activity of beclin-1 wild-type 3′-UTR construct in U87 cells. However, the mutated (MUT) construct of beclin-1 3′-UTR significantly abolished this inhibitory effect of miR-17-5p in U87 cells (Fig. 5B). Our results suggested that beclin-1 was the direct target of miR-17-5p.
Figure 5.
miR-17-5p directly targets 3′-UTR of beclin-1. (A) The predicted binding sequences for beclin-1 3′-UTR with miR-17-5p. (B) The interaction between miR-17-5p and beclin-1 was determined by luciferase activity assays. The wild-type or mutant beclin-1 3′-UTR was cotransfected with miR-17-5p mimics or scramble miRNA and incubated for 48 h into U-87 cells followed with a detection by dual-luciferase reporter assay kit. WT denotes pGL3 vectors containing beclin-1 3′-UTR; MUT denotes pGL3 vectors containing mutated beclin-1 3′-UTR. **p < 0.01 compared with the indicated group.
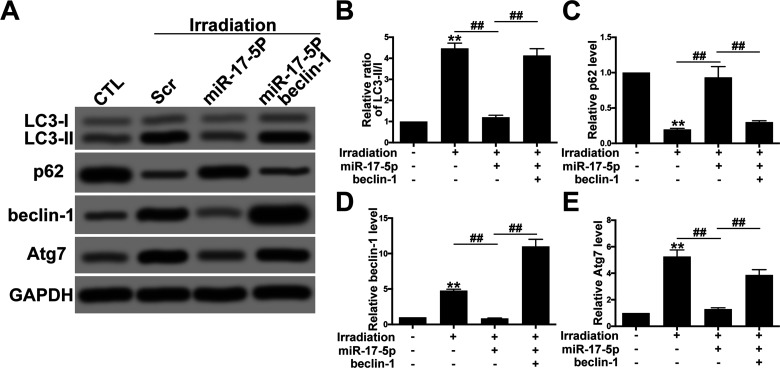
Exotic Expression of miR-17-5p Decreased Autophagy Activity via Targeting Beclin-1
To further explore the potential relationship between miR-17-5p and autophagy in RR-U87 cells, we transfected RR-U87 cells with miR-17-5p mimics and determined the alteration of autophagy. Western blot showed that miR-17-5p significantly decreased beclin-1, the ratio of LC3-II/I as well as Atg7 levels and augmented p62 levels, suggesting that miR-17-5p played an important role in inhibiting irradiation-induced autophagy activity (Fig. 6A–E). However, overexpression of beclin-1 obviously reversed the inhibitory effect of miR-17-5P on autophagy activity.
Figure 6.
Exotic expression of miR-17-5p decreased autophagy activity via targeting beclin-1. (A) U87 cells were transfected with miR-17-5p mimics with or without beclin-1 overexpression plasmids. Then the cells were subjected to irradiation, and the autophagy activity was tested by Western blot. (B–E) Relative levels of ratio of LC3-II/I (B), p62 (C), beclin-1 (D), and Atg7 (E) were measured using ImageJ and normalized to GAPDH. Data are presented as mean ± SD from at least three independent experiments. **p < 0.01 compared with control group and ##p < 0.01 compared with the indicated groups.
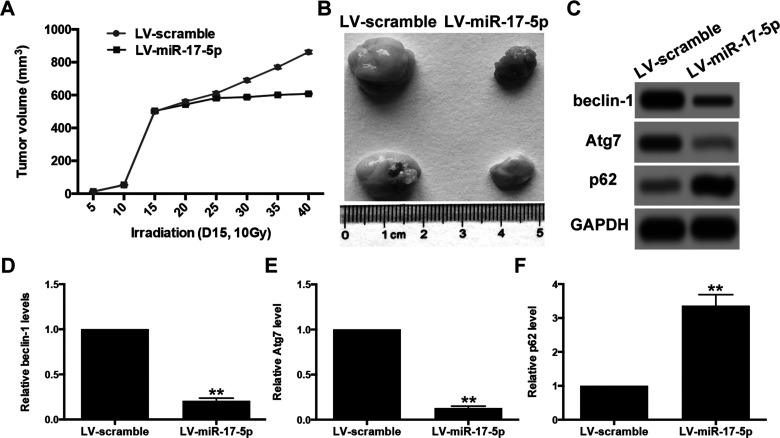
miR-17-5p Sensitized Xenograft Tumor to Irradiation and Inhibited Irradiation-Induced Autophagy
To determine whether miR-17-5p can increase the efficiency of irradiation in killing implanted tumors in nude mice, we subcutaneously injected nude mice with U87 cells pretransfected with lentiviral vector expressing miR-17-5p or scramble miRNA control. When the tumors reached about 500 mm3 on day 15, a single dose of 10 Gy irradiation was given. The results showed that miR-17-5p significantly increased the radiosensitivity of U87-derived tumors to irradiation treatment and markedly reduced the tumor volume, compared with the scramble group (Fig. 7A and B). We further analyzed the levels of beclin-1 and autophagy activity in the xenograft tumor after irradiation. Western blot showed that miR-17-5p inhibited irradiation-induced autophagy activation as indicated by the downregulation of beclin-1 and Atg7 as well as the upregulation of p62 (Fig. 7C–F). These in vivo findings confirmed that miR-17-5p sensitized glioma cells to irradiation by blocking irradiation-induced beclin-1-mediated autophagy.
Figure 7.
miR-17-5p sensitized xenograft tumor to irradiation and inhibited irradiation-induced autophagy. (A) Nude mice were subcutaneously injected with U87 cells infected with control lentivirus vector or miR-129-5p overexpression lentivirus vector. The tumors were irradiated (10 Gy) on day 15 when the average tumor volume reached about 500 mm3. Tumor volume was monitored over time as indicated. (B) The tumors were excised on day 40. miR-129-5p overexpression caused a significant decrease in tumor volume. (C–F) the autophagy activity was determined by Western blot for beclin-1, Atg7, and p62. Relative density of protein bands was measured using ImageJ and normalized to GAPDH. Data are presented as mean ± SD from at least three independent experiments. **p < 0.01 compared with LV-scramble group.
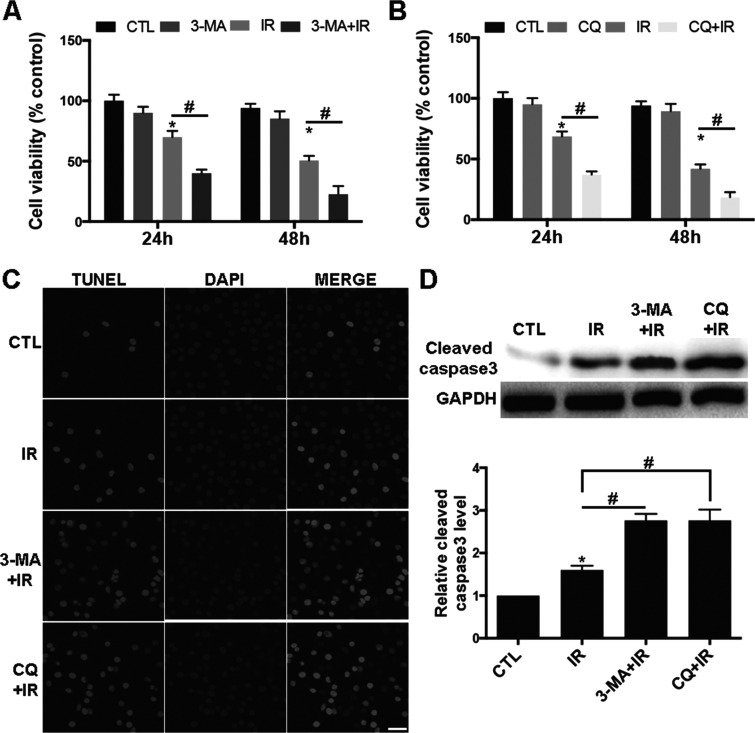
Pharmacal Inhibition of Autophagy Sensitized RR-U87 Cells to Irradiation
We next applied the autophagy inhibitors 3-MA and CQ to investigate the role of autophagy in the radiosensitivity of glioma cells. 3-MA is a specific inhibitor of autophagosome formation, whereas CQ suppresses lysosomal acidification and degradation (21). RR-U87 cells were pretreated with 4 μM 3-MA or 20 μM CQ for 1 h and then exposed to irradiation (2 Gy/min). The cytotoxic effect of irradiation was measured by MTT. Compared with the control group, 3-MA or CQ pretreatment significantly sensitized RR-U87 cells to irradiation-induced cell death (Fig. 8A and B). In addition, 3-MA or CQ pretreatment markedly increased apoptosis in RR-U87 cells as indicated by TUNEL assay (Fig. 8C) and Western blot for cleaved caspase 3 (Fig. 8D). These data suggested that pharmacal inhibition of autophagy sensitized RR-U87 cells to irradiation.
Figure 8.
Pharmacal inhibition of autophagy sensitized RR-U87 cells to irradiation. (A, B) RR-U87 cells were pretreated with 4 μM 3-MA or 20 μM CQ for 1 h and then exposed to irradiation (2 Gy/min). The cell viability at 24 and 48 h after irradiation was measured by MTT. Data are presented as percentage normalized to control group and are presented as mean ± SD from at least three independent experiments. CTL, control; IR, irradiation. Compared with the control group, 3-MA or CQ pretreatment significantly sensitized RR-U87 cells to irradiation-induced cell death. (C) TUNEL assay was conducted to detect the apoptosis in RR-U87 cells after autophagy was suppressed with 3-MA or CQ . Scale bar: 50 μm. (D) Cleaved caspase 3 was detected with Western blot, and relative level of cleaved caspase 3 was measured using ImageJ and normalized to GAPDH. Data are presented as mean ± SD from at least three independent experiments. *p < 0.05 compared with control group and #p < 0.05 compared with the indicated groups.
DISCUSSION
In the present study, we provided evidence that six miRNAs were downregulated and seven were upregulated in the serum after radiotherapy compared with paired serum samples before radiotherapy via miRNA panel PCR. Among these, miR-17-5p was highly reduced (fold change = −4.21). Further, we validated the levels of miR-17-5p in all serum samples with qRT-PCR. In addition, statistical analysis suggested that a reduced miR-17-5P level was positively associated with the advanced clinical stage of glioma, incidence of relapse, and tumor differentiation. Moreover, we provided evidence that irradiation markedly decreased miR-17-5p and activated autophagy in the glioma cell line as evidenced by the upregulation of ratio of LC-3II, Atg7, and beclin-1 as well as downregulation of autophagic target protein p62. Pharmacal inhibition of autophagy markedly enhanced the cytotoxicity of irradiation in RR-U87 cells. Further, we demonstrated that irradiation-induced autophagy activation was mediated by beclin-1, and downregulation of beclin-1 via siRNA significantly abolished the irradiation-activated autophagy. Interestingly, we demonstrated that beclin-1 is a direct target of miR-17-5p via luciferase gene reporter assay. Exotic expression of miRNA-17-5p decreased autophagy activity in vitro. Finally, in nude mice, miRNA-17-5p sensitized xenograft tumor to irradiation and inhibited irradiation-induced autophagy.
miRNAs have been characterized as important regulatory mechanisms of gene expression and intensively studied in tumorigenesis and cancer treatment. miR-17-5p could regulate the progression of hepatocellular carcinoma through targeting PTEN (22). Also, many other studies show that miR-17-5p plays an important antitumor role in diffuse large B-cell lymphoma, cervical cancer cells, breast cancer cells, and ovarian cancer cells (23–26). However, studies on the role of miR-17-5p in radiosensitivity of glioma cells are still lacking. In this study, our results demonstrated that miR-17-5p significantly enhanced the radiosensitivity of glioma cells and robustly inhibits the proliferation and survival of glioma cells. Importantly, we demonstrated that the action of miR-17-5p in glioma cells was associated with inhibiting beclin-1-mediated autophagy, suggesting beclin-1 as a potential target in clinical treatment for glioma. In addition, our present study screened out six decreased and seven increased miRNAs in the serum after irradiation compared with the paired serum samples before irradiation. Among these, miR-126 acts as a tumor suppressor in glioma cells by targeting insulin receptor substrate 1 (IRS-1) (27). miR-24 regulates the proliferation and invasion of glioma by ST7L through the β-catenin pathway (28). miR-134 modulates proliferation and invasion of glioma cells by targeting KRAS and suppressing the ERK pathway (29). Downregulating miR-155 induces apoptosis and enhances chemosensitivity of U251 cells to taxol (30). miR-335 is a potential biomarker for poor prognosis in glioma (31). miR-503 regulates L1CAM to suppress proliferation and invasion of glioma cells (32). However, their relationship with the radiosensitivity of glioma cells is still completely unknown, which warrants further studies.
Autophagy, an adaptive response against cellular stress, has been shown to be involved in the radioresistance of cancer cells (33,34). In glioma, radiation induces autophagy, which is responsible for the radioresistance of glioma stem cells (35,36). In breast cancer, inhibition of autophagy promoted radiosensitivity via inhibiting transforming growth factor-activated kinase-1 (37). In addition, hypoxia-induced autophagy was demonstrated to contribute to the radioresistance of breast cancer cells (38). The role of autophagy in the regulation of radioresistance of pancreatic cancer was also investigated. Profilin1 is capable of sensitizing pancreatic cancer cells to irradiation by suppressing autophagy (39). As expected, the present study demonstrated that irradiation significantly activated autophagy and provided direct evidence that autophagy played a vital role in irradiation-associated glioma cellular death with autophagy inhibitor 3-MA and CQ. Our study reinforced the evidence for the role of autophagy in regulating radioresistance of cancer cells. Previous studies demonstrated that autophagy could be mediated by CDK5, HO-1 or beclin-1, etc., under different stimuli or diseases (40–42). Herein, our study confirmed the vitally mediative role of beclin-1 in irradiation-induced autophagy in U87 cells, suggesting beclin-1 as a potential target in clinical treatment for glioma. However, it is still essential to explore the roles of other autophagic mediators, such as CDK5 and HO-1, in further studies.
In conclusion, the present study provides evidence that miR-17-5p plays an important role in radiosensitivity of glioma cells by inhibiting beclin-1-mediated autophagy. Targeting miR-17-5p/beclin-1/autophagy pathway may be a promising strategy to enhance the efficacy of radiotherapy in glioma.
ACKNOWLEDGMENTS
The authors appreciate Prof. Simon from the Department of Neurology, Massachusetts General Hospital, Boston, USA, for the critical reading of the manuscript.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Schwartzbaum J. A.; Fisher J. L.; Aldape K. D.; Wrensch M. Epidemiology and molecular pathology of glioma. Nat. Clin. Pract. Neurol. 2:494–503; 2006. [DOI] [PubMed] [Google Scholar]
- 2. Henke G.; Paulsen F.; Steinbach J. P.; Ganswindt U.; Isijanov H.; Kortmann R. D.; Bamberg M.; Belka C. Hypofractionated reirradiation for recurrent malignant glioma. Strahlenther Onkol. 185:113–119; 2009. [DOI] [PubMed] [Google Scholar]
- 3. Narayana A.; Yamada J.; Berry S.; Shah P.; Hunt M.; Gutin P. H.; Leibel S. A. Intensity-modulated radiotherapy in high-grade gliomas: Clinical and dosimetric results. Int. J. Radiat. Oncol. Biol. Phys. 64:892–897; 2006. [DOI] [PubMed] [Google Scholar]
- 4. Douw L.; Klein M.; Fagel S. S.; van den Heuvel J.; Taphoorn M. J.; Aaronson N. K.; Postma T. J.; Vandertop W. P.; Mooij J. J.; Boerman R. H.; Beute G. N.; Sluimer J. D.; Slotman B. J.; Reijneveld J. C.; Heimans J. J. Cognitive and radiological effects of radiotherapy in patients with low-grade glioma: Long-term follow-up. Lancet Neurol. 8:810–818; 2009. [DOI] [PubMed] [Google Scholar]
- 5. Frosina G. DNA repair and resistance of gliomas to chemotherapy and radiotherapy. Mol. Cancer Res. 7:989–999; 2009. [DOI] [PubMed] [Google Scholar]
- 6. Bao S.; Wu Q.; McLendon R. E.; Hao Y.; Shi Q.; Hjelmeland A. B.; Dewhirst M. W.; Bigner D. D.; Rich J. N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444:756–760; 2006. [DOI] [PubMed] [Google Scholar]
- 7. Filipowicz W.; Bhattacharyya S. N.; Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 9:102–114; 2008. [DOI] [PubMed] [Google Scholar]
- 8. Selbach M.; Schwanhausser B.; Thierfelder N.; Fang Z.; Khanin R.; Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature 455:58–63; 2008. [DOI] [PubMed] [Google Scholar]
- 9. Song L.; Liu S.; Zhang L.; Yao H.; Gao F.; Xu D.; Li Q. MiR-21 modulates radiosensitivity of cervical cancer through inhibiting autophagy via the PTEN/Akt/HIF-1alpha feedback loop and the Akt-mTOR signaling pathway. Tumour Biol.; 2016. [E-pub ahead of print] [DOI] [PubMed] [Google Scholar]
- 10. Mao A.; Zhao Q.; Zhou X.; Sun C.; Si J.; Zhou R.; Gan L.; Zhang H. MicroRNA-449a enhances radiosensitivity by downregulation of c-Myc in prostate cancer cells. Sci. Rep. 6:27346; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang R.; Ye F.; Zhen Q.; Song T.; Tan G.; Chu W.; Zhang Y.; Lv B.; Zhao X.; Liu J. MicroRNA-148b is a potential prognostic biomarker and predictor of response to radiotherapy in non-small-cell lung cancer. J. Physiol. Biochem. 72:337–343; 2016. [DOI] [PubMed] [Google Scholar]
- 12. Yen W. L.; Klionsky D. J. How to live long and prosper: Autophagy, mitochondria, and aging. Physiology (Bethesda) 23:248–262; 2008. [DOI] [PubMed] [Google Scholar]
- 13. Mijaljica D.; Prescott M.; Devenish R. J. Endoplasmic reticulum and Golgi complex: Contributions to, and turnover by, autophagy. Traffic 7:1590–1595; 2006. [DOI] [PubMed] [Google Scholar]
- 14. Wang B.; Cai Z.; Tao K.; Zeng W.; Lu F.; Yang R.; Feng D.; Gao G.; Yang Q. Essential control of mitochondrial morphology and function by chaperone-mediated autophagy through degradation of PARK7. Autophagy 12:1–14; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kroemer G.; Marino G.; Levine B. Autophagy and the integrated stress response. Mol. Cell 40:280–293; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang P.; Zhang J.; Zhang L.; Zhu Z.; Fan J.; Chen L.; Zhuang L.; Luo J.; Chen H.; Liu L.; Chen Z.; Meng Z. MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology 145:1133–1143 e1112; 2013. [DOI] [PubMed] [Google Scholar]
- 17. Yang Y.; Yang Y.; Yang X.; Zhu H.; Guo Q.; Chen X.; Zhang H.; Cheng H.; Sun X. Autophagy and its function in radiosensitivity. Tumour Biol. 36:4079–4087; 2015. [DOI] [PubMed] [Google Scholar]
- 18. Huang Q.; Zhan L.; Cao H.; Li J.; Lyu Y.; Guo X.; Zhang J.; Ji L.; Ren T.; An J.; Liu B.; Nie Y.; Xing J. Increased mitochondrial fission promotes autophagy and hepatocellular carcinoma cell survival through the ROS-modulated coordinated regulation of the NFKB and TP53 pathways. Autophagy 1–16; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nam H. Y.; Han M. W.; Chang H. W.; Kim S. Y.; Kim S. W. Prolonged autophagy by MTOR inhibitor leads radioresistant cancer cells into senescence. Autophagy 9:1631–1632; 2013. [DOI] [PubMed] [Google Scholar]
- 20. Wang B.; Cai Z.; Lu F.; Li C.; Zhu X.; Su L.; Gao G.; Yang Q. Destabilization of survival factor MEF2D mRNA by neurotoxin in models of Parkinson’s disease. J. Neurochem. 130:720–728; 2014. [DOI] [PubMed] [Google Scholar]
- 21. Mizushima N.; Yoshimori T.; Levine B. Methods in mammalian autophagy research. Cell 140:313–326; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shan S. W.; Fang L.; Shatseva T.; Rutnam Z. J.; Yang X.; Du W.; Lu W. Y.; Xuan J. W.; Deng Z.; Yang B. B. Mature miR-17-5p and passenger miR-17-3p induce hepatocellular carcinoma by targeting PTEN, GalNT7 and vimentin in different signal pathways. J. Cell. Sci. 126:1517–1530; 2013. [DOI] [PubMed] [Google Scholar]
- 23. Robertus J. L.; Harms G.; Blokzijl T.; Booman M.; de Jong D.; van Imhoff G.; Rosati S.; Schuuring E.; Kluin P.; van den Berg A. Specific expression of miR-17-5p and miR-127 in testicular and central nervous system diffuse large B-cell lymphoma. Mod. Pathol. 22:547–555; 2009. [DOI] [PubMed] [Google Scholar]
- 24. Wei Q.; Li Y. X.; Liu M.; Li X.; Tang H. MiR-17-5p targets TP53INP1 and regulates cell proliferation and apoptosis of cervical cancer cells. IUBMB Life 64:697–704; 2012. [DOI] [PubMed] [Google Scholar]
- 25. Taylor D. D.; Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110:13–21; 2008. [DOI] [PubMed] [Google Scholar]
- 26. Hossain A.; Kuo M. T.; Saunders G. F. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol. Cell. Biol. 26:8191–8201; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luan Y.; Zuo L.; Zhang S.; Wang G.; Peng T. MicroRNA-126 acts as a tumor suppressor in glioma cells by targeting insulin receptor substrate 1 (IRS-1). Int. J. Clin. Exp. Pathol. 8:10345–10354; 2015. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28. Chen L.; Zhang A.; Li Y.; Zhang K.; Han L.; Du W.; Yan W.; Li R.; Wang Y.; Wang K.; Pu P.; Jiang T.; Jiang C.; Kang C. MiR-24 regulates the proliferation and invasion of glioma by ST7L via beta-catenin/Tcf-4 signaling. Cancer Lett. 329:174–180; 2013. [DOI] [PubMed] [Google Scholar]
- 29. Zhao Y.; Pang D.; Wang C.; Zhong S.; Wang S. MicroRNA-134 modulates glioma cell U251 proliferation and invasion by targeting KRAS and suppressing the ERK pathway. Tumour Biol.; 2016. [E-pub ahead of print] [DOI] [PubMed] [Google Scholar]
- 30. Meng W.; Jiang L.; Lu L.; Hu H.; Yu H.; Ding D.; Xiao K.; Zheng W.; Guo H.; Ma W. Anti-miR-155 oligonucleotide enhances chemosensitivity of U251 cell to taxol by inducing apoptosis. Cell Biol. Int. 36:653–659; 2012. [DOI] [PubMed] [Google Scholar]
- 31. Jiang J.; Sun X.; Wang W.; Jin X.; Bo X.; Li Z.; Bian A.; Jiu J.; Wang X.; Liu D.; Hui X.; Wang Y.; Wang A.; Ding L. Tumor microRNA-335 expression is associated with poor prognosis in human glioma. Med. Oncol. 29:3472–3477; 2012. [DOI] [PubMed] [Google Scholar]
- 32. Liu H.; Song Z.; Liao D.; Zhang T.; Liu F.; Zheng W.; Luo K.; Yang L. miR-503 inhibits cell proliferation and invasion in glioma by targeting L1CAM. Int. J. Clin. Exp. Med. 8:18441–18447; 2015. [PMC free article] [PubMed] [Google Scholar]
- 33. Chaachouay H.; Ohneseit P.; Toulany M.; Kehlbach R.; Multhoff G.; Rodemann H. P. Autophagy contributes to resistance of tumor cells to ionizing radiation. Radiother. Oncol. 99:287–292; 2011. [DOI] [PubMed] [Google Scholar]
- 34. Zois C. E.; Koukourakis M. I. Radiation-induced autophagy in normal and cancer cells: Towards novel cytoprotection and radio-sensitization policies? Autophagy 5:442–450; 2009. [DOI] [PubMed] [Google Scholar]
- 35. Lomonaco S. L.; Finniss S.; Xiang C.; Decarvalho A.; Umansky F.; Kalkanis S. N.; Mikkelsen T.; Brodie C. The induction of autophagy by gamma-radiation contributes to the radioresistance of glioma stem cells. Int. J. Cancer 125:717–722; 2009. [DOI] [PubMed] [Google Scholar]
- 36. Apel A.; Herr I.; Schwarz H.; Rodemann H. P.; Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 68:1485–1494; 2008. [DOI] [PubMed] [Google Scholar]
- 37. Han M. W.; Lee J. C.; Choi J. Y.; Kim G. C.; Chang H. W.; Nam H. Y.; Kim S. W.; Kim S. Y. Autophagy inhibition can overcome radioresistance in breast cancer cells through suppression of TAK1 activation. Anticancer Res. 34:1449–1455; 2014. [PubMed] [Google Scholar]
- 38. He W. S.; Dai X. F.; Jin M.; Liu C. W.; Rent J. H. Hypoxia-induced autophagy confers resistance of breast cancer cells to ionizing radiation. Oncol. Res. 20:251–258; 2012. [DOI] [PubMed] [Google Scholar]
- 39. Cheng H.; Li J.; Liu C.; Yao W.; Xu Y.; Frank T. S.; Cai X.; Shi S.; Lu Y.; Qin Y.; Liu L.; Xu J.; Long J.; Ni Q. X.; Li M.; Yu X. J. Profilin1 sensitizes pancreatic cancer cells to irradiation by inducing apoptosis and reducing autophagy. Curr. Mol. Med. 13:1368–1375; 2013. [DOI] [PubMed] [Google Scholar]
- 40. Wong A. S.; Lee R. H.; Cheung A. Y.; Yeung P. K.; Chung S. K.; Cheung Z. H.; Ip N. Y. Cdk5-mediated phosphorylation of endophilin B1 is required for induced autophagy in models of Parkinson’s disease. Nat. Cell Biol. 13:568–579; 2011. [DOI] [PubMed] [Google Scholar]
- 41. Carchman E. H.; Rao J.; Loughran P. A.; Rosengart M. R.; Zuckerbraun B. S. Heme oxygenase-1-mediated autophagy protects against hepatocyte cell death and hepatic injury from infection/sepsis in mice. Hepatology 53:2053–2062; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matsui Y.; Takagi H.; Qu X.; Abdellatif M.; Sakoda H.; Asano T.; Levine B.; Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: Roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ. Res. 100:914–922; 2007. [DOI] [PubMed] [Google Scholar]