Abstract
miRNAs have been shown to be involved in breast cancer growth and progression. miR-4262 is a potential tumor promoter in human cancers. In this study, we first investigated the role of miR-4262 in the proliferation and invasion of human breast cancer cells. Our results showed that, compared with the adjacent tissues and MCF-10A normal breast epithelial cells, miR-4262 was markedly increased in the breast cancer tissues and five cell lines, including MDA-MB-231, MDA-MB-468, MDA-MB-435, SKBR3, and MCF-7. Then the miR-4262 mimic or oligo anta-miR-4262 was transfected into MDA-MB-231 and MCF-7 breast cancer cell lines. The results showed that the miR-4262 mimic greatly increased the miR-4262 level and the proliferation and invasion of MDA-MB-231 and MCF-7 cells. In contrast, the anta-miR-4262 had a completely opposite effect on miR-4262 expression, cell proliferation, and cell invasion in MDA-MB-231 and MCF-7 cells. Moreover, bioinformatics and luciferase reporter gene assays confirmed that miR-4262 targeted the mRNA 3′-UTR region of KLF6 and KLF15, two characterized tumor suppressor genes. miR-4262 suppressed protein levels of KLF6 and KLF15 in MDA-MB-231 cells, and the suppression could be rescued by the transfection of pcDNA-KLF6 and -KLF15. In conclusion, miR-4262 positively regulates proliferation and invasion of human breast cancer cells via suppression of KLF6 and KLF15.
Key words: miR-4262, Breast cancer, Cell proliferation and invasion, KLF6, KLF15
INTRODUCTION
Breast cancer is the leading tumor threat to women’s physical and mental health. Its incidence ranks first among malignant tumors in females, and the survival rate of patients is relatively low (1). In recent years, drugs that inhibit the invasion and metastasis of breast cancer are constantly emerging. However, the death rate of breast cancer patients is still very high in developing countries (2,3). Therefore, it is urgent to improve the existing treatment strategies and develop novel therapeutic targets.
Mammary glands are not required for basic life activities in humans, so breast cancer in situ is not so life threatening. However, once the metastatic breast cancer invades into other tissues/organs, the survival rate of patients will drop sharply (4). The invasion and metastasis of breast cancer involve activation of many oncogenes, suppression of tumor suppressor genes, and dysregulation of some signaling pathways (4,5). miRNAs are small noncoding RNAs that suppress the expression of specific genes by targeting their 3′-untranslated regions (3′-UTRs) (6). They play important roles in diverse biological processes by targeting various genes, including normal tissue development and tumor progression (6,7). A number of miRNAs have been demonstrated to play a critical role in breast cancer progression (8,9). For example, let-7 miRNAs were markedly reduced in self-renewing, tumor-initiating, and differentiated cells from breast cancer lines, and they could inhibit proliferation and promote apoptosis of breast cancer cells (10).
miR-4262 is a newly found miRNA that was first shown to be associated with cisplatin resistance in adenocarcinoma cells in 2013 (11). From then on, several large-scale miRNA expression profiles showed that miR-4262 potentially played a promotive role in several human diseases, such as alcoholic cardiomyopathy and acute lung injury (12,13). Quite recently, miR-4262 was found to regulate osteosarcoma (OS) cell invasion through osteopontin modification (14). However, the exact role of miR-4262 in the growth and progression of breast cancer is completely unknown.
In this study, the miR-4262 expression was detected in breast cancer and normal adjacent tissues, as well as in human normal breast epithelial cells and breast cancer cell lines. We found that miR-4262 was distinctly upregulated in cancerous tissues and cell lines. Oligo miR-4262 mimic and anta-miR-4262 were, respectively, transfected into MDA-MB-231 and MCF-7 human breast cancer cell lines. We first report that miR-4262 plays a positive role in breast cancer cell proliferation and invasion. Furthermore, the underlying mechanism was explored.
MATERIALS AND METHODS
Ethical Statements
This study was approved by the ethics committee of the First Affiliated Hospital of Xi’an Jiaotong University (P.R. China). The subjects were informed of experimental details and gave written consent.
Tissue Sampling
Breast cancer and matched normal adjacent tissues were isolated from a total of 48 breast cancer patients (54 ± 6.8 years) who underwent tumor resection during the last 2 years in our hospital. Patients who ever received chemo- or radiotherapy or suffered acute lung/heart/kidney diseases were excluded from this study.
Cell Culture and Transfection
Human normal breast epithelial cell line MCF-10A and human breast cancer cell lines, including MDA-MB-231, MDA-MB-468, MDA-MB-435, SKBR3, and MCF-7, and HEK293 cells, were purchased from the American Type Culture Collection Company (ATCC; Manassas, VA, USA). They were cultured in RPMI-1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco), 100 U/ml penicillin (Gibco), and 100 U/ml streptomycin (Gibco). The cells were incubated at a humidified atmosphere at 37°C in a 5% CO2 incubator.
For transfection, MDA-MB-231 and MCF-7 breast cancer cells were subcultured in six-well plates. On reaching 70% confluence, 60 nM miR-4262 mimic oligo, 60 nM miR-4262 antagomir oligo, and 60 nM relative control oligos were transfected into the cells by using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. For validating the effect of miR-4262 on the expression of KLF6 and KLF15, 60 nM miR-4262 mimic was transfected alone or cotransfected with 4 μg of pcDNA-KLF6/KLF15 (2 μg of pcDNA-KLF6 and 2 μg of pcDNA-KLF15) into MDA-MB-231 cells. The miR-4262 mimic, anta-miR-4262, and their control oligos were designed and synthesized by the Ribobio Co., Ltd. (Guangzhou, P.R. China). The pcDNA-KLF6 and pcDNA-KLF15 were presented by Dr. X. B. Wang (College of Animal Science, Northwest A&F University).
Cell Proliferation
The cells were seeded in six-well culture plates at 105 cells/per well. After transfection for 48 h, cell proliferation was evaluated using the cell counting kit-8 (CCK-8; Sigma-Aldrich, St. Louis, MO, USA) assay according to the manufacturer’s instructions.
In Vitro Invasion Assay
Transwell invasion assay was used to detect cell invasion. Cells (3.5 × 104) with serum-free medium were plated in the top chamber of the plates (24-well insert; 8 μm; Corning). Medium containing 10% serum was used as a chemoattractant in the lower chamber. After incubation for 24 h, a cotton swab was used to remove the nonmigrated cells in the upper chamber, and the filters were individually stained with 2% crystal violet. The invasive cells were captured under a light microscope (100×; Olympus IX70; Olympus Corporation, Osaka, Japan) and counted.
Real-Time Quantitative PCR
Real-time qPCRs were carried on in a 25-μl system using SYBR Premix Ex Taq (Invitrogen), 0.4 mM primers, and 200 ng of cDNA template. The primers of miR-4262 and U6 RNA (internal reference) were designed and produced by Ribobio Co., Ltd. PCR amplification cycles were performed using the SYBR Premix Ex Taq II Kit (Invitrogen). The reactions were initially denatured at 95°C for 1 min and followed by 35 cycles of 94°C for 15 s and 60°C for 1 min. The transcript abundance was calculated using the 2−ΔΔCt method.
Western Blotting
Total protein (30 μg) from each sample was separated by 12% SDS-PAGE gel. The protein was transferred onto a PVDF membrane (Millipore, Billerica, MA, USA). The primary antibodies KLF6 (1:300), KLF15 (1:300), EGFR (1:400), pEGFR (1:200), CCND1 (1:400), and β-actin (1:800) (all from Abcam) were used to incubate with the membrane at 4°C overnight. After incubation with the HRP-conjugated secondary antibody for 1 h at room temperature, proteins were detected with an ECL Kit (Millipore) and analyzed with a UV Transilluminator (Bio-Rad).
3′-UTR Luciferase Gene Reporter Assay
The 3′-UTR sequences of human KLF6 and KLF15 mRNAs were amplified by PCR. The sequences were, respectively, purified and inserted into a psiCHECK™-2 Vector (Promega, Madison, WI, USA). The control mimic and miR-4262 mimic were, respectively, cotransfected with psiCHECK-KLF6 or KLF15 3′-UTR into the HEK293 cells using Lipofectamine 3000 (Invitrogen). The cells were then incubated for 72 h, and luciferase activity was measured using a microplate reader (PT-3502; Potenov, Beijing, P.R. China).
Statistical Analysis
All measurements were obtained from more than three independent experiments. The data were expressed as means ± SEM. Statistics were calculated with SPSS v23.0. Multiple comparisons were assessed by one-way ANOVA followed by Dunnett’s tests. A value of p < 0.05 was considered statistically significant.
RESULTS
miR-4262 Was Distinctly Upregulated in Human Breast Cancer Tissues and Cell Lines
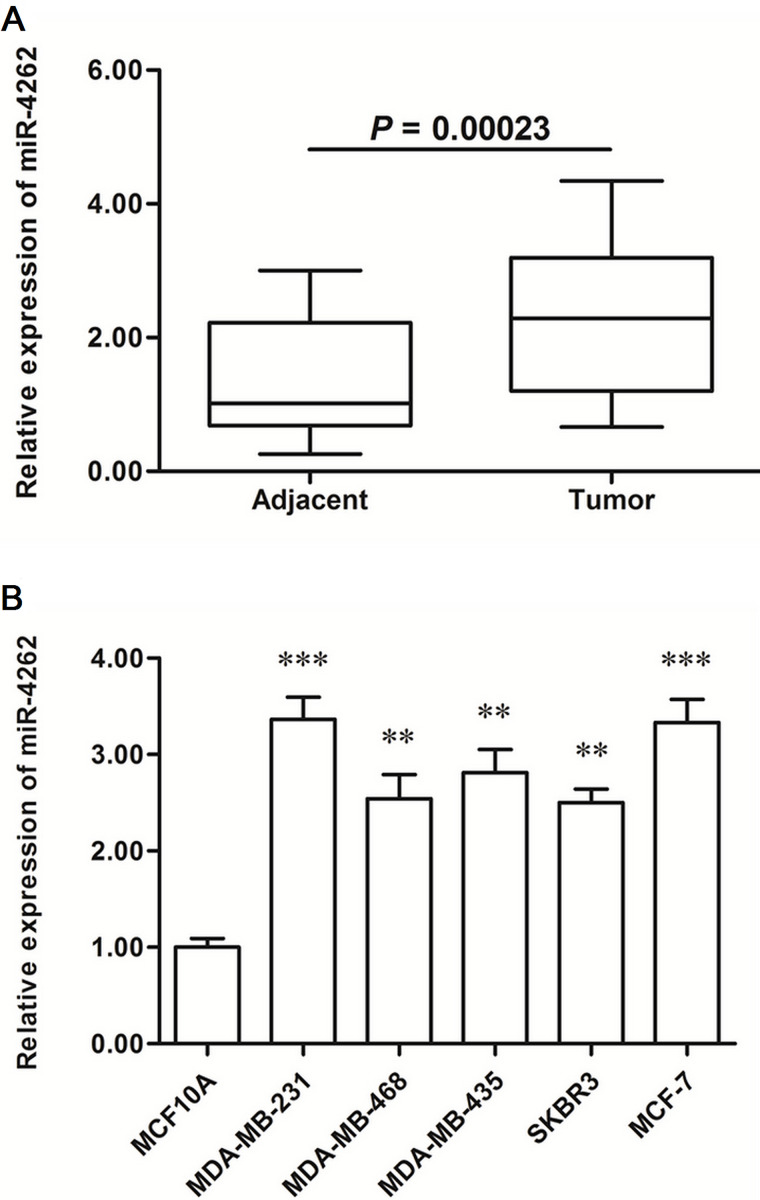
The expression levels were detected in breast cancer tissues and matched adjacent normal tissues. Real-time qPCR analysis showed that miR-4262 was increased by about twofold in the cancerous tissues (p < 0.0001) (Fig. 1A). Then the miR-4262 levels in human normal breast epithelial cell line and five breast cancer cell lines were also detected. The results showed that miR-4262 was increased by 2.5-fold in all cancer cell lines (p < 0.01), even to threefold in MDA-MB-231 and MCF-7 cells (p < 0.001) (Fig. 1B).
Figure 1.
miR-4262 was significantly upregulated in human breast cancer tissues and cell lines. Breast cancer tissues and paired adjacent noncancerous tissues were sampled from 48 breast cancer patients. The expression levels of miR-4262 were detected in these tissues with real-time qPCR. Then the expression levels of miR-4262 were detected in the human normal breast epithelial cell MCF-10A and five human breast cancer cell lines including MDA-MB-231, MDA-MB-468, MDA-MB-435, SKBR3, and MCF-7. (A) miR-4262 was significantly upregulated in breast cancer tissues. (B) miR-4262 was significantly elevated in breast cancer cell lines. **p < 0.01, ***p < 0.001 versus MCF-10A.
miR-4262 Mimic Transfection Increased Proliferation and Invasion of Human Breast Cancer Cells
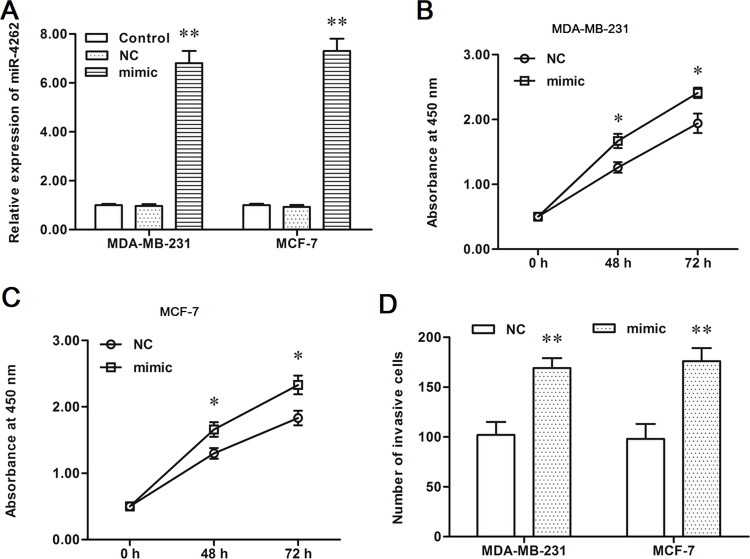
To investigate the role of miR-4262 in the progression of human breast cancer, a negative control mimic (NC) and miR-4262 mimic were, respectively, transfected into MDA-MB-231 and MCF-7 breast cancer cells. After incubation for 48 h, miR-4262 expression, cell proliferation, and cell invasion were detected. The data on real-time qPCR analysis showed that miR-4262 expression was increased by about sevenfold by the miR-4262 mimic transfection in both MDA-MB-231 and MCF-7 cells (p < 0.01) (Fig. 2A). CCK-8 and Transwell invasion assays showed that the miR-4262 mimic increased proliferation and invasion of MDA-MB-231 and MCF-7 cells (p < 0.05) (Fig. 2B–D).
Figure 2.
miR-4262 mimic transfection promoted proliferation and invasion of MDA-MB-231 and MCF-7 breast cancer cell lines. The negative control mimic (NC) or miR-4262 mimic was transfected into MDA-MB-231 and MCF-7 human breast cancer cell lines. After incubation for 48 h, miR-4262 expression was detected with real-time qPCR, cell proliferation was detected with the CCK-8 method, and cell invasion was detected with the Transwell invasion assay. (A) miR-4262 expression was increased greatly by the miR-4262 mimic transfection. (B, C) The miR-4262 mimic significantly promoted the proliferation of MDA-MB-231 and MCF-7 cells. (D) The miR-4262 mimic significantly enhanced the invasion of MDA-MB-231 and MCF-7 cells. *p < 0.05, *p < 0.01 versus NC.
miR-4262 Inhibition Suppressed Proliferation and Invasion of MDA-MB-231 and MCF-7 Cells
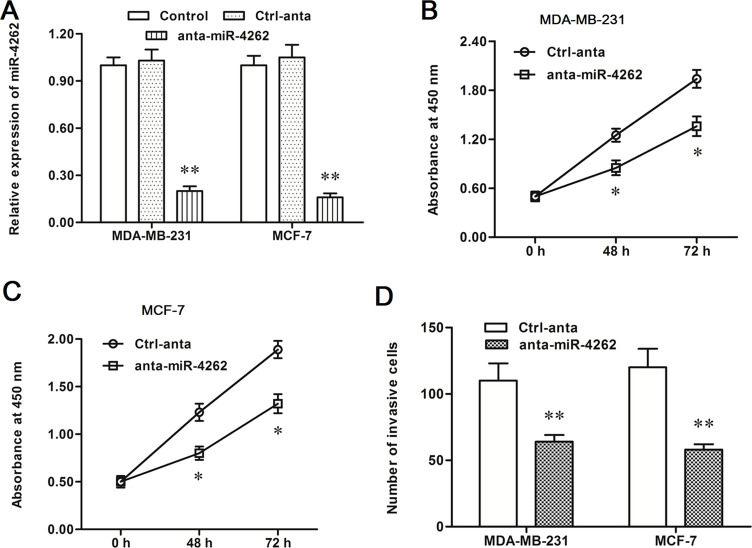
Subsequently, the control anta oligo and anta-miR-4262 oligo were transfected into MDA-MB-231 and MCF-7 cells, respectively. In contrast to the results from the miR-4262 mimic transfection, miR-4262 expression was decreased by about 80% in both MDA-MB-231 and MCF-7 cells (p < 0.01) (Fig. 3A). The miR-4262 inhibitor sharply suppressed proliferation and invasion of MDA-MB-231 and MCF-7 cells (p < 0.05) (Fig. 3B–D). The results from the mimic and inhibitor transfection indicated that miR-4262 positively regulated proliferation and invasion of breast cancer cells.
Figure 3.
miR-4262 antagomir increased proliferation and invasion of MDA-MB-231 and MCF-7 cells. The control or antagomir miR-4262 oligo was transfected into MDA-MB-231 and MCF-7 cells. After incubation for 48 h, miR-4262 expression, cell proliferation, and cell invasion were detected. (A) The miR-4262 level was sharply decreased by the anta-miR-4262 transfection. (B, C) The anta-miR-4262 suppressed the proliferation of MDA-MB-231 and MCF-7 cells. (D) The anta-miR-4262 suppressed the invasion of MDA-MB-231 and MCF-7 cells. *p < 0.05, **p < 0.01 versus Ctrl-anta.
miR-4262 Directly Targeted the Tumor Suppressor Genes KLF6 and KLF15 to Suppress Their Expression
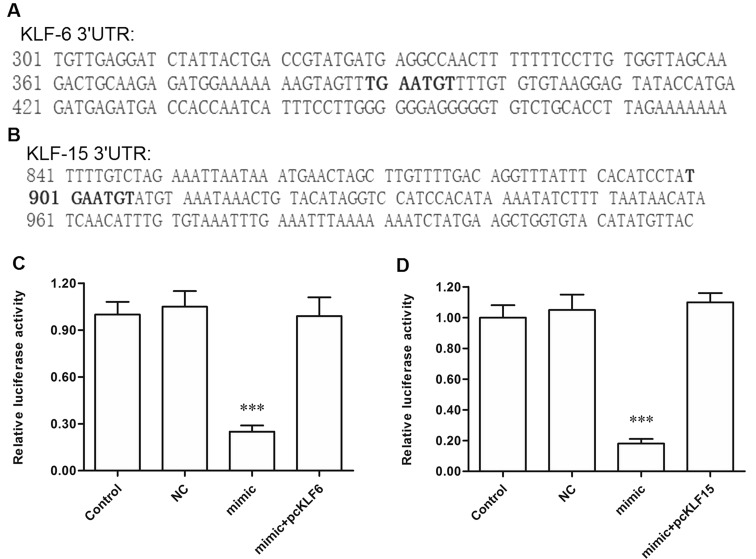
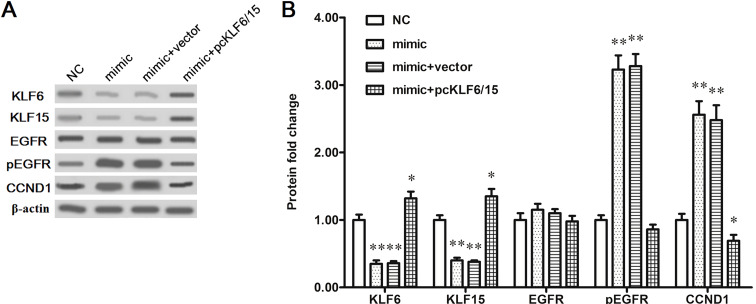
To explore the potential mechanism of miR-4262 promoting proliferation and invasion of breast cancer cells, bioinformatics analysis was then used to seek its potential targets. The output of miRDB showed that the 3′-UTR region of seven consecutive bases in KLF6 and KLF15 mRNA sequences completely matched with the seed of miR-4262 (Fig. 4A and B). The 3′-UTR luciferase reporter gene assay showed that miR-4262 markedly suppressed the luciferase activity, and pcDNA-KLF6 or pcDNA-KLF15 could rescue the suppression (Fig. 4C and D). To further verify that KLF6 and KLF15 were target genes of miR-4262, the NC or miR-4262 mimic was transfected alone or cotransfected with pcDNA-KLF6 or pcDNA-KLF15 into MDA-MB-231 cells. Western blotting indicated that the KLF6 and KLF15 protein levels were sharply reduced by the miR-4262 mimic transfection, and the reduction could be reversed by the pcDNA-KLF6/KLF15 transfection (Fig. 5A and B). These results indicated that miR-4262 directly targets the tumor suppressor genes KLF6 and KLF15.
Figure 4.
miR-4262 targeted KLF6 and KLF15 mRNA at the 3′-UTR region. (A, B) Outputs of the miRDB online servers on the binding of miR-4262 with the 3′-UTR region of KLF6 and KLF15 mRNAs. (C, D) Validation of the binding of miR-4262 with KLF6 and KLF15 mRNAs by the 3′-UTR luciferase reporter gene assay. ***p < 0.001 versus control.
Figure 5.
miR-4262 negatively regulated the protein levels of KLF6 and KLF15. The MDA-MB-231 was transfected with 60 nM NC mimic, 60 nM miR-4262 mimic, 60 nM miR-4262 mimic plus 2 μg of pcDNA3.1 empty vector, or 60 nM miR-4262 mimic plus 2 μg of pcDNA-KLF6 and 2 μg of pcDNA-KLF15. After incubation for 48 h, Western blotting was used to detect the protein levels of KLF6, KLF15, EGFR, pEGFR, and CCND1. (A) The blots of KLF6, KLF15, EGFR, pEGFR, and CCND1. (B) Statistical analysis for (A). *p < 0.05, **p < 0.01 versus NC.
DISCUSSION
miR-4262 is a newly found miRNA and has been poorly characterized to date. In this study, we report that miR-4262 expression was distinctly increased in human breast cancer tissues and cell lines, compared with normal adjacent tissues and normal breast epithelial cells, suggesting a positive role in the progression of breast cancer. Our data on the miR-4262 mimic and antagomir oligo transfection showed that miR-4262 had a promotive effect on the proliferation and invasion of breast cancer cells. This is the first report, to our knowledge, that indicated the role of miR-4262 in the progression of human breast cancer.
In fact, there have been reports revealing the association of miR-4262 with a couple of cancers. One of the reports showed that miR-4262 levels were significantly decreased in OS specimens, compared with the paired adjacent nontumor tissue; miR-4262 overexpression inhibited osteopontin-mediated cell invasion; and miR-4262 depletion increased osteopontin-mediated cell invasion in OS cells (14). Moreover, the 5-year survival of patients with lower miR-4262 levels in the resected OS was worse than that of patients with high miR-4262 levels (14). However, other research showed that miR-4262 enhanced the proliferation of hepatocellular carcinoma cells and played a promotive role in hepatocellular carcinoma progression (15). The role of miR-4262 seems to be contradictory in diverse cancers. In this study, we found that miR-4262 plays a promotive role in the proliferation and invasion of breast cancer cells.
The Krüppel-like family of transcription factors (KLFs) are a set of zinc finger DNA-binding proteins, of which the biological roles of the KLFs are of wide interest (16). KLF6 is an identified tumor suppressor gene in multiple cancers (17–20). Some research showed that downregulation of KLF6 mRNA levels was described in lung and esophageal cancers (21,22). Other research indicated that reduced KLF6 expression levels in lung and prostate cancers have been reported to be correlated with increased likelihood of recurrence (23,24). Activation of KLF6 triggered transactivation of p21 and EGFR and suppressed cyclin D1 (CCND1)-induced cell proliferation and EGFR-induced cell invasion/metastasis (25–27). KLF15 has recently been implicated in the development of several human malignancies, including breast cancer (28). It was indicated that exogenous KLF15 overexpression resulted in a concomitant decrease in cellular proliferation and an arrest at the G0/G1 phase of the cell cycle. KLF15 upregulation could result in activation of p21, a pivotal suppressor of the G1/S phase transition, and suppress CCND1-mediated cell proliferation (29). Several miRNAs have been shown to play a positive role in cancer growth and metastasis by targeting KLF15 or KLF6. For instance, miRNA-181a promotes gastric cancer by negatively regulating KLF6 (30). In this study, our bioinformatics analysis and 3′-UTR luciferase reporter gene assay showed that miR-4262 directly targeted the 3′-UTR region of KLF6 and KLF15 mRNA sequences and negatively regulated their expression.
In conclusion, miR-4262 expression was upregulated in human breast cancer tissues and cell lines. miR-4262 had a promotive effect on the proliferation and invasion of breast cancer cells by directly targeting the tumor suppressor genes KLF6 and KLF15.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Goldhirsch A.; Winer E. P.; Coates A.; Gelber R.; Piccart-Gebhart M.; Thürlimann B.; Senn H.-J.; Albain K. S.; André F.; Bergh J. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 24:2206–2223; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. DeSantis C.; Ma J.; Bryan L.; Jemal A. Breast cancer statistics, 2013. CA Cancer J. Clin. 64:52–62; 2014. [DOI] [PubMed] [Google Scholar]
- 3. Siegel R.; Naishadham D.; Jemal A. Cancer statistics, 2013. CA Cancer J. Clin. 63:11–30; 2013. [DOI] [PubMed] [Google Scholar]
- 4. Stevanovic A.; Lee P.; Wilcken N. Metastatic breast cancer. Aust. Fam. Physician 35:309; 2006. [PubMed] [Google Scholar]
- 5. Cristofanilli M.; Budd G. T.; Ellis M. J.; Stopeck A.; Matera J.; Miller M. C.; Reuben J. M.; Doyle G. V.; Allard W. J.; Terstappen L. W. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 351:781–791; 2004. [DOI] [PubMed] [Google Scholar]
- 6. Croce C. M.; Calin G. A. miRNAs, cancer, and stem cell division. Cell 122:6–7; 2005. [DOI] [PubMed] [Google Scholar]
- 7. Valencia-Sanchez M. A.; Liu J.; Hannon G. J.; Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20:515–524; 2006. [DOI] [PubMed] [Google Scholar]
- 8. Iorio M. V.; Ferracin M.; Liu C.-G.; Veronese A.; Spizzo R.; Sabbioni S.; Magri E.; Pedriali M.; Fabbri M.; Campiglio M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 65:7065–7070; 2005. [DOI] [PubMed] [Google Scholar]
- 9. Tavazoie S. F.; Alarcón C.; Oskarsson T.; Padua D.; Wang Q.; Bos P. D.; Gerald W. L.; Massagué J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451:147–152; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yu F.; Yao H.; Zhu P.; Zhang X.; Pan Q.; Gong C.; Huang Y.; Hu X.; Su F.; Lieberman J. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131:1109–1123; 2007. [DOI] [PubMed] [Google Scholar]
- 11. Pouliot L. M.; Shen D.-W.; Suzuki T.; Hall M. D.; Gottesman M. M. Contributions of microRNA dysregulation to cisplatin resistance in adenocarcinoma cells. Exp. Cell Res. 319:566–574; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bao H.; Gao F.; Xie G.; Liu Z. Angiotensin-converting enzyme 2 inhibits apoptosis of pulmonary endothelial cells during acute lung injury through suppressing mir-4262. Cell. Physiol. Biochem. 37:759–767; 2015. [DOI] [PubMed] [Google Scholar]
- 13. Jing L.; Jin C.; Lu Y.; Huo P.; Zhou L.; Wang Y.; Tian Y. Investigation of microRNA expression profiles associated with human alcoholic cardiomyopathy. Cardiology 130:223–233; 2015. [DOI] [PubMed] [Google Scholar]
- 14. Song K.; Liu N.; Yang Y.; Qiu X. Regulation of osteosarcoma cell invasion through osteopontin modification by miR-4262. Tumor Biol. 1–7; 2015. [DOI] [PubMed] [Google Scholar]
- 15. Lu S.; Wu J.; Gao Y.; Han G.; Ding W.; Huang X. MicroRNA-4262 activates the NF-κB and enhances the proliferation of hepatocellular carcinoma cells. Int. J. Biol. Macromol. 86:43–49; 2016. [DOI] [PubMed] [Google Scholar]
- 16. Pearson R.; Fleetwood J.; Eaton S.; Crossley M.; Bao S. Krüppel-like transcription factors: A functional family. Int. J. Biochem. Cell B 40:1996–2001; 2008. [DOI] [PubMed] [Google Scholar]
- 17. Narla G.; Heath K. E.; Reeves H. L.; Li D.; Giono L. E.; Kimmelman A. C.; Glucksman M. J.; Narla J.; Eng F. J.; Chan A. M. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science 294:2563–2566; 2001. [DOI] [PubMed] [Google Scholar]
- 18. Chen C.; Hyytinen E.-R.; Sun X.; Helin H. J.; Koivisto P. A.; Frierson H. F.; Vessella R. L.; Dong J.-T. Deletion, mutation, and loss of expression of KLF6 in human prostate cancer. Am. J. Pathol. 162:1349–1354; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reeves H. L.; Narla G.; Ogunbiyi O.; Haq A. I.; Katz A.; Benzeno S.; Hod E.; Harpaz N.; Goldberg S.; Tal-Kremer S. Kruppel-like factor 6 (KLF6) is a tumor-suppressor gene frequently inactivated in colorectal cancer. Gastroenterology 126:1090–1103; 2004. [DOI] [PubMed] [Google Scholar]
- 20. Kremer-Tal S.; Reeves H. L.; Narla G.; Thung S. N.; Schwartz M.; Difeo A.; Katz A.; Bruix J.; Bioulac-Sage P.; Martignetti J. A. Frequent inactivation of the tumor suppressor Kruppel-like factor 6 (KLF6) in hepatocellular carcinoma. Hepatology 40:1047–1052; 2004. [DOI] [PubMed] [Google Scholar]
- 21. Tahara E.; Kadara H.; Lacroix L.; Lotan D.; Lotan R. Activation of protein kinase C by phorbol 12-myristate 13-acetate suppresses the growth of lung cancer cells through KLF6 induction. Cancer Biol. Ther. 8:801–807; 2009. [DOI] [PubMed] [Google Scholar]
- 22. Tetreault M.-P.; Yang Y.; Katz J. P. Kruppel-like factors in cancer. Nat. Rev. Cancer 13:701–713; 2013. [DOI] [PubMed] [Google Scholar]
- 23. Ito G.; Uchiyama M.; Kondo M.; Mori S.; Usami N.; Maeda O.; Kawabe T.; Hasegawa Y.; Shimokata K.; Sekido Y. Krüppel-like factor 6 is frequently down-regulated and induces apoptosis in non-small cell lung cancer cells. Cancer Res. 64:3838–3843; 2004. [DOI] [PubMed] [Google Scholar]
- 24. Chiam K.; Ryan N. K.; Ricciardelli C.; Day T. K.; Buchanan G.; Ochnik A. M.; Murti K.; Selth L. A.; Butler L. M.; Tilley W. D. Characterization of the prostate cancer susceptibility gene KLF6 in human and mouse prostate cancers. Prostate 73:182–193; 2013. [DOI] [PubMed] [Google Scholar]
- 25. Sangodkar J.; Dhawan N.; Melville H.; Singh V. J.; Farrington C.; Yuan E.; Rana H.; Smith B.; Gidwani V.; Okrent R. Targeting the FOXO1/KLF6 transcriptional network to modulate response to anti-EGFR based therapy. Cancer Res. 72:1885–1885; 2012. [Google Scholar]
- 26. Lang U.; Kocabayoglu P.; Cheng G.; Ghiassi-Nejad Z.; Muñoz U.; Vetter D.; Eckstein D.; Hannivoort R.; Walsh M.; Friedman S. GSK3β phosphorylation of the KLF6 tumor suppressor promotes its transactivation of p21. Oncogene 32:4557–4564; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benzeno S.; Narla G.; Allina J.; Cheng G. Z.; Reeves H. L.; Banck M. S.; Odin J. A.; Diehl J. A.; Germain D.; Friedman S. L. Cyclin-dependent kinase inhibition by the KLF6 tumor suppressor protein through interaction with cyclin D1. Cancer Res. 64:3885–3891; 2004. [DOI] [PubMed] [Google Scholar]
- 28. Yoda T.; McNamara K. M.; Miki Y.; Onodera Y.; Takagi K.; Nakamura Y.; Ishida T.; Suzuki T.; Ohuchi N.; Sasano H. KLF15 in breast cancer: A novel tumor suppressor? Cell. Oncol. 38:227–235; 2015. [DOI] [PubMed] [Google Scholar]
- 29. Hong Q.; Li C.; Xie Y.; Lv Y.; Liu X.; Shi S.; Ding R.; Zhang X.; Zhang L.; Liu S. Kruppel-like factor-15 inhibits the proliferation of mesangial cells. Cell. Physiol. Biochem. 29:893–904; 2012. [DOI] [PubMed] [Google Scholar]
- 30. Zhang X.; Nie Y.; Du Y.; Cao J.; Shen B.; Li Y. MicroRNA-181a promotes gastric cancer by negatively regulating tumor suppressor KLF6. Tumor Biol. 33:1589–1597; 2012. [DOI] [PubMed] [Google Scholar]