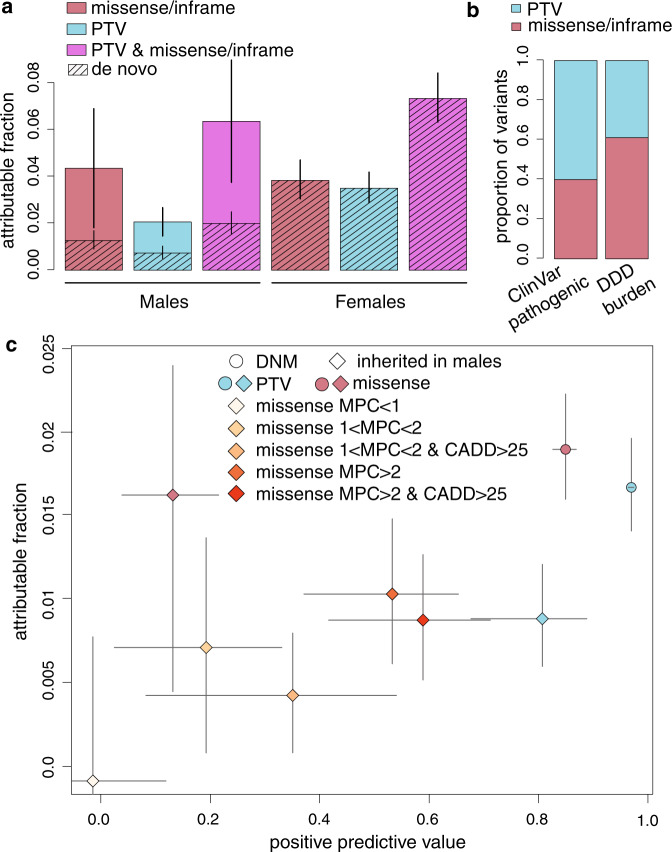
Fig. 1. Results from burden analysis of rare and de novo coding variants on the X chromosome.
a Fraction of males and females attributable to rare inherited and de novo coding variants on the X chromosome. Note that in the males, the overall attributable fraction was estimated from the case/control analysis of all male probands (7136 cases versus 8551 controls), whereas that for de novo mutations (DNMs) was estimated only in the 5138 male trio probands. In the females, only de novos were considered since we were assuming full penetrance. b Relative fraction of protein-truncating variants (PTVs) versus missense/inframe variants amongst ClinVar likely pathogenic or pathogenic variants in X-linked DDG2P genes, versus the fraction inferred in the burden analysis in DDD. c Estimated attributable fraction versus positive predictive value for DNMs and inherited variants in males in X-linked DDG2P genes. Inherited missense variants are split according to CADD (Combined Annotation Dependent Depletion) 21 and MPC (missense badness, PolyPhen, constraint) 22 scores. In a, c the coloured bars (a) or points (c) show the point estimate and the error bars show 95% confidence intervals calculated as described in the ‘Methods’.