Table 1.
Employable imaging techniques for the investigation of TME interactions in zebrafish.
| Technique | Applications | Penetration | Disadvantages | References | Example |
|---|---|---|---|---|---|
| Stereomicroscopy | Possesses potential for fluorescent and time-lapse imaging (live and fixed) | Not limited | Requires transparent fish | Paatero et al., 2018 | 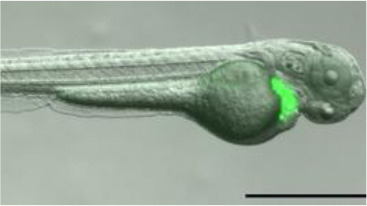 |
| Conventional confocal microscopy | 3D imaging and time lapses (live and fixed) | Up to 200 μm | Can be time-consuming | van den Berg et al., 2019 | 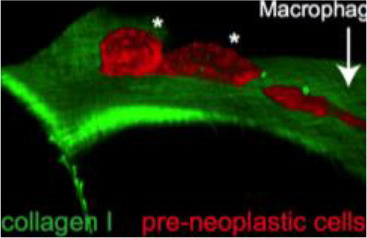 |
| Correlative light and electron microscopy | Multimodal: 3D imaging with definition of ultrastructure (live and fixed) | Up to 200 μm | Time-consuming | van den Berg et al., 2019 | 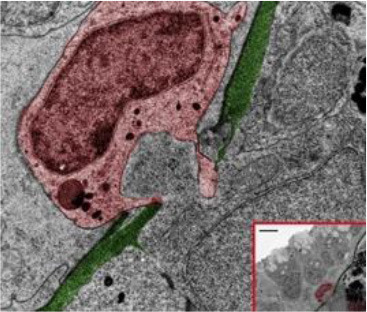 |
| Two photon (multiphoton) | Cellular behavior and membrane order; commonly use fluorescent dyes or endogenous markers (live and fixed) | Up to 500 μm | Potential for thermal damage; decreased molecular brightness | Perrin et al., 2020 | 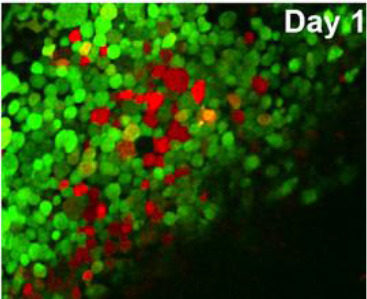 |
| Second harmonic generation (multiphoton) | Non-centrosymmetric structures like collagen fibers (live and fixed) | Up to 300 μm | Limited applicability to structural proteins | LeBert et al., 2016 | 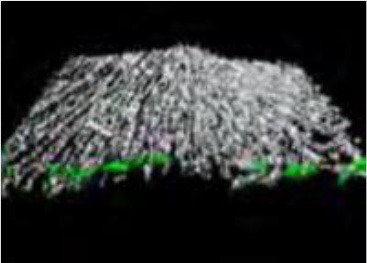 |
| Selective plane illumination microscopy (light sheet) | 3D imaging, deep optical sectioning (live and fixed) | Up to 3 mm | Extra optics required | Gualda et al., 2015 | 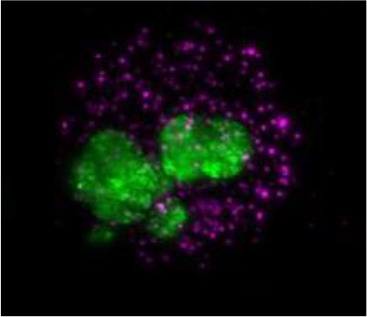 |
| Micro-CT | 3D whole-organism imaging; phenotypic and architectural (live and fixed) | Not limited | Time-consuming | Ding et al., 2019 | 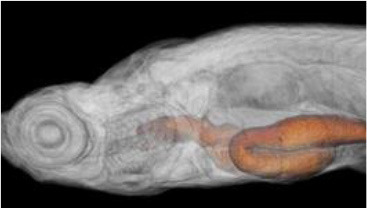 |
TME, tumor microenvironment.
