Figure 1.
Design of RNA recovery strategy for cell-specific biopanning
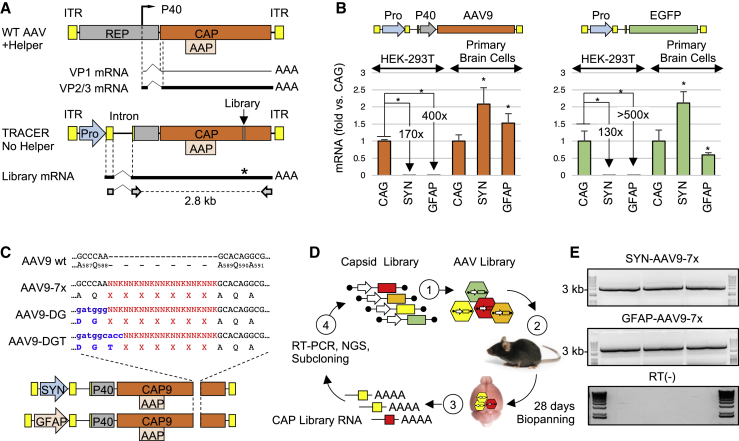
(A) Map of wild-type AAV (top) and TRACER library vectors (bottom). ITR, inverted terminal repeat. Pro, promoter. Dashed lines indicate AAV intron (top) or synthetic CMV-globin intron (bottom), solid lines represent minor (thin line) and major (thick line) capsid transcripts. Primers used for the recovery of the 2.8-kb capsid library amplicon are indicated at the bottom. (B) Activity of CAG, SYN, and GFAP promoters in TRACER tandem configuration (left panel) or single promoter configuration (right panel). Depicted transgenes were packaged in AAV9 capsid and tested in HEK293T cells or primary mouse brain cells (1e5 VG/cell, n = 3). RNA was quantified by real-time RT-PCR 48 h post-treatment. Values indicate RNA expression normalized to CAG vectors in each cell type (mean ± SD). ∗p < 0.05 (t test). (C) Construction of peptide display libraries. Randomized sequences preceded by AQ, DG, or DGT residues were introduced in AAV9 VP1 at the indicated positions in vectors containing SYN or GFAP promoter. (D) Overview of the in vivo selection process. (1) DNA libraries are used to produce a virus library, (2) virus libraries are injected intravenously (i.v.) into mice (1e12VG per mouse), (3) bulk RNA is recovered from whole brains 28 days post-injection, (4) capsid fragments encoding the peptide library are amplified by RT-PCR, analyzed by next-generation sequencing (NGS), and cloned into TRACER vectors for another round of selection. (E) Example of RT-PCR products obtained from three mice 28 days after injection with SYN-driven and GFAP-driven library (top and middle panel, respectively). The 3-kb band from the molecular weight marker is indicated. Bottom panel: RT-negative controls.