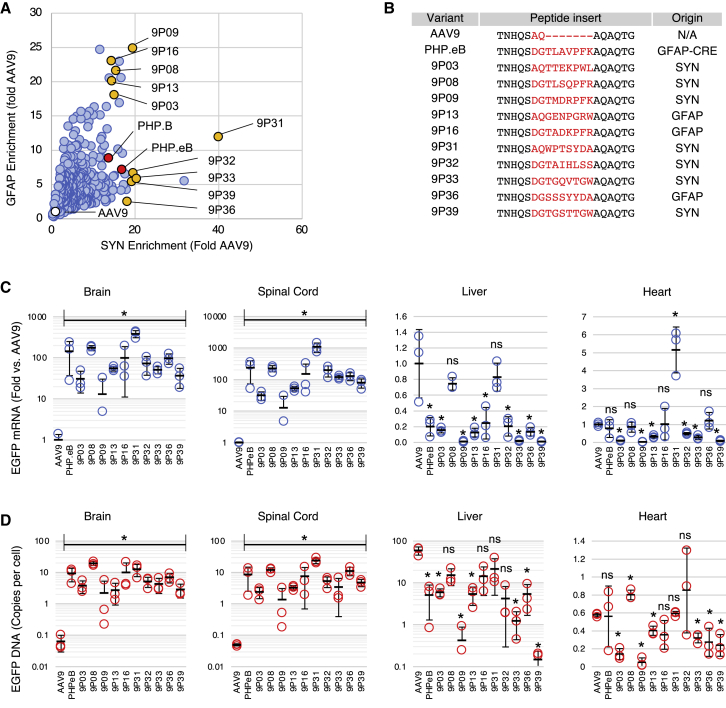
Figure 4.
Individual characterization of TRACER capsid candidates
(A) Brain RNA enrichment score of pooled synthetic variants in the SYN- and GFAP-driven assay. Values are normalized to AAV9. Yellow dots indicate candidates chosen for individual testing; red and white dots indicate the PHP and AAV9 control capsids, respectively. (B) Sequence of capsids selected for individual characterization. The right column indicates the biopanning method used to evolve each variant. (C) Real-time RT-PCR analysis of EGFP transgene RNA expression in the brain, spinal cord, liver, and heart 28 days after i.v. injection of each capsid in C57BL/6 mice (4e11 VG per mouse, n = 3). Values indicate mean ± SD (n = 3), normalized to AAV9. ∗p < 0.05 relative to AAV9 (unpaired t test); ns, not significant. (D) AAV genome biodistribution measured by qPCR. Values indicate mean ± SD (n = 3) EGFP copies per diploid cell. ∗p < 0.05 relative to AAV9 (unpaired t test); ns, not significant. All brain and spinal cord samples were statistically different from AAV9.