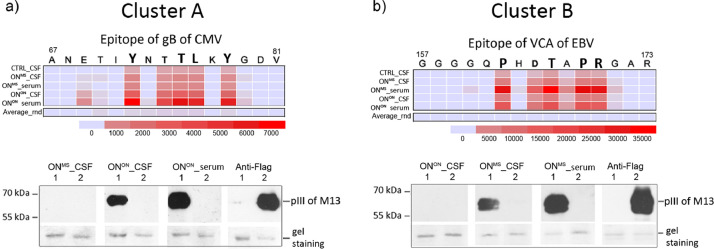
Fig. 3.
Epitope clusters A and B mimic highly antigenic epitopes of CMV and EBV. a-b) Heatmap images of epitopes of clusters A and B alignments to IEDB epitopes (01.08.2019). The criteria for homology searches were set to 4 amino acid similarity matches and group median values for each epitope motif were used in alignment calculations. On top of the heatmap panels primary sequences of gB CMV (Uniprot code Q2FAM8) and VCA p18 EBV (Uniprot code P14348) are shown with amino acids defining the core epitopes of clusters A and B. Below the heatmap panels, the scale of relative alignment loads (0–35 000) is shown in colour code. The acronyms of the discovery and validation cohorts are shown in the left. Average_rnd – average median values of scrambled motifs derived of clusters A and B motifs. Representative images from validation studies of antigenic epitope predictions using western blot analysis are shown in the lower part of the figure (see full blots on Figure S5). Western blot analysis of recombinant phages containing the epitope gB CMV or VCA EBV was performed using recombinant phages encoding Y..TL.Y-pIII or P..T.PR-pIII (1) and flag-pIII fusion proteins (2) with primary antibodies: i) pre-cleared serum/plasma (dilution 1:750), CSF (dilution 1:7,5); ii) mouse anti-FLAG antibody (Sigma Aldrich, No. 287) and secondary antibodies: i) rabbit anti-human-HRP (Abcam), ii) rabbit anti-mouse-HRP (Abcam). Protein molecular weight markers (kDa) are shown in the left. Gel staining – protein loading control with Coomassie blue staining, pIII – pIII protein of M13. Source of primary antibodies is indicated on top of the blot.