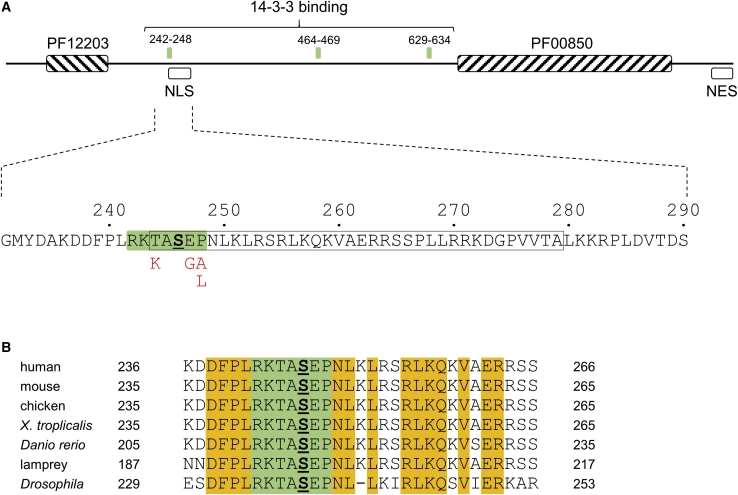
Figure 1.
Schematic organization of HDAC4 and location of variants
(A) The upper figure shows the organization of the 1,084-residue HDAC4 protein; hatched boxes show Pfam domains PF12203 (glutamine-rich N-terminal domain of HDAC4; residues 62–152) and PF00850 (histone deacetylase domain; residues 675–992) as indicated; green boxes indicate known sites of 14-3-3 binding; open boxes show positions of the nuclear localization signal (NLS) and nuclear export signal (NES). The region around the first 14-3-3 site is shown expanded in the lower figure; the 14-3-3 site spanning residues 242–248 is shaded green, with the phosphorylated serine, Ser246, underlined in bold font; the NLS (244–279) is boxed; missense variants described in this report (p.Thr244Lys, p.Glu247Gly, p.Pro248Ala, and p.Pro248Leu) are shown below the HDAC4 sequence in red font.
(B) Residues 236–266 of HDAC4 are shown aligned to orthologs; the core 14-3-3 site, RKTASEP, is shaded green and is invariant in all sequences; other invariant residues are shaded orange. UniProtKB accession codes for HDAC4 orthologs are as follows: human, UniProtKB: P56524; mouse, UniProtKB: Q6NZM9; chicken, UniProtKB: P83038; Xenopus tropicalis, UniProtKB: F7CSW6; Danio rerio, UniProtKB: Q08BS8; lamprey, UniProtKB: S4RJL9; Drosophila melanogaster, UniProtKB: Q9VYF3.