Figure 3.
In silico analysis of HDAC4 variants
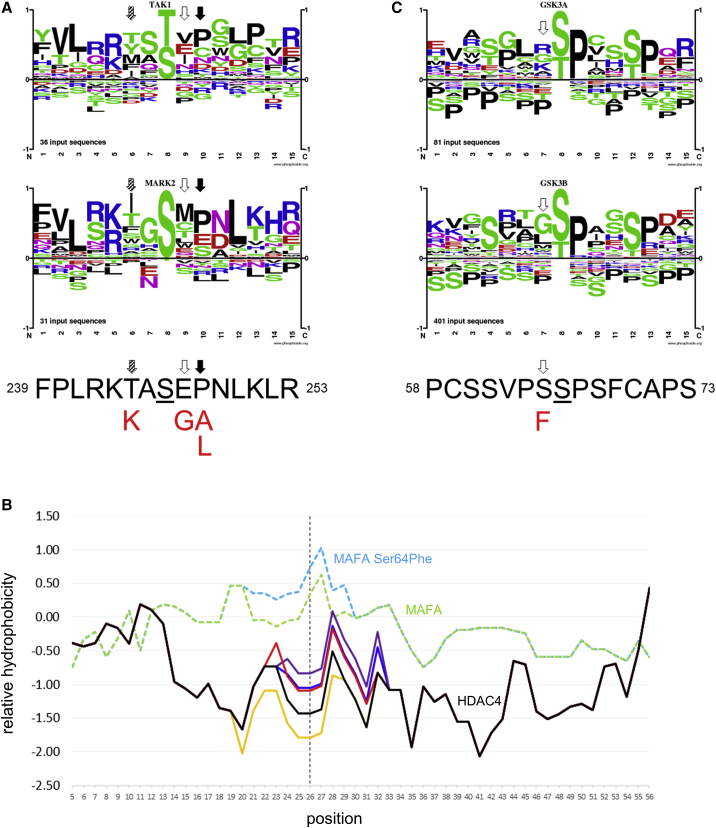
(A) Upper panels show sequence logos indicating substrate specificity of TAK1 and MARK2 as indicated; color indicates sidechain properties (blue, positive; red, negative; magenta, neutral; green, polar; black, hydrophobic), and all sequences are centered on the phosphorylated serine or threonine residue at position 8 of the logo. Below these is shown the sequence of HDAC4 residues 239–253 (black font; the phosphorylated serine, Ser246, is underlined); residues Glu247 and Pro248 are indicated by open and filled arrows, respectively, and these align to positions 9 and 10 of substrate logos (or +1 and +2 relative to the phosphorylated serine) as marked; residue Thr244 is marked by a hatched arrow and lies at position 6 of the logo (−2 relative to Ser246); variants observed at these positions are shown in red font below the sequence.
(B) Relative hydrophobicity is shown for residues 221–280 of HDAC4 (solid black line) and for variants p.Glu247Gly, p.Pro248Ala, p.Pro248Leu, and p.Thr244Lys (solid red, blue, purple, and gold lines, respectively); broken lines show hydrophobicity of residues 20–79 of MAFA (green) and MAFA variant Ser64Phe (light blue); in all cases, traces are aligned to show the phosphorylated residue (HDAC4 Ser246; MAFA Ser65) at position 26 of the analysis, as indicated by the vertical broken line.
(C) Similar to (A), showing substrate specificity logos for GSK3A (top) and GSK3B (center); the sequence below shows residues 58–73 of MAFA, with the phosphorylated serine (Ser65) underlined; the position of the p.Ser64Phe variant is shown by an open arrow in all parts.