Abstract
The interaction between the host plant Arabidopsis thaliana (Arabidopsis) and the oomycete Hyaloperonospora arabidopsidis (Hpa) is an established model system for the study of an obligate biotrophic downy mildew interaction. The evaluation of the developmental success of Hpa is often based on the quantification of reproductive structures that are formed on the surface of leaves, such as the sporangiophores or the conidiospores they carry. However, the structural basis of this interaction lies within the plant tissue and, in particular, the haustoria that form inside plant cells. Therefore, valuable additional information about the performance and compatibility of the downy mildew interaction can be gained by light microscopical inspection of the hyphal and haustorial shape inside the plant tissue and within plant cells respectively. Here we describe a protocol for the visualization and quantification of morphological phenotypes inside the plant. While we focus specifically on the quantification of haustorial shape variants, the protocol can easily be adapted for the quantification of other morphological features such as hyphal deformations, or oogonia frequency. By including and refining already existing protocols from a variety of sources, we assembled the entire experimental pipeline for the Arabidopsis Hpa bioassay to provide a practical guide for the initial setup of this system in the laboratory. This pipeline includes the following steps: A) growing Arabidopsis, B) Hpa propagation and strain maintainance C) Hpa inoculation and incubation D) staining of plant tissues for visualization of the pathogen and E) an introduction of the Keyence VHX microscope and Fiji plugin of ImageJ for the quantification of structures of interest. While described here for Arabidopsis and Hpa, the protocol steps B-E should be easily adjustable for the study of other plant-oomycete pathosystems.
Keywords: Arabidopsis, Hyaloperonospora arabidopsidis, Trypan blue staining, Haustoria quantification, Multilobed haustoria, Plant-microbe interaction
Background
Downy mildews are a group of phytopathogenic oomycetes characterized by branched sporangiophores that protrude through leaf stomata, giving the appearance of a white to grey “down” on the affected tissues (Agrios, 2005). Hundreds of Downy mildew species have been described on a wide variety of host plants including monocotyledon and dicotyledon crops (Callan and Carris, 2004). They are typically obligate biotrophs, meaning that they have not yet been successfully cultured outside the plant host with completion of their life cycle. A model system to study the molecular and genetic determinants underlying the downy mildew disease is the interaction between the host plant Arabidopsis thaliana and its oomycetal pathogen Hyaloperonospora arabidopsidis (Hpa). Downy mildew oomycetes typically form a structure called haustorium inside the plant host cell. The haustorium is surrounded by a plant cell membrane called extrahaustorial membrane (EHM) that prevents direct contact of the oomycete with the plant cytoplasm. While the hypothetical role of the haustorium in nutrient uptake from the plant host remains to be confirmed, it probably represents a platform for delivering oomycetal effectors to plants (Judelson and Ah-Fong, 2019) which enable the establishment and maintenance of a biotrophic interaction with the host. It is therefore not surprising that several proteins required for the resistance against Hpa are localized to the EHM ( Wang et al., 2009 ; Caillaud et al., 2014 ). Despite the obvious relevance of the haustorium for the downy mildew interaction, surprisingly few studies evaluated the interaction by phenotyping the haustoria. A method for the inoculation of Arabidopsis by Hpa was described by Asai et al. (2015) . Here we expanded this protocol resulting in a step-by-step guide for the study of this pathosystem in the lab. This includes details on how to sow and grow Arabidopsis, how to propagate Hpa, and specific details on how to systematically record structural features of this interaction with the Keyence VHX digital microscope, and quantify them with the image analysis software Fiji, here exemplified by the quantification of haustoria-shape variation. Our step-by-step guide for the microscopic analysis of the Hpa-Arabidopsis interaction should be easily adaptable for the use not only in other downy mildew interactions in host species other than Arabidopsis but also other hyphal pathogens other than oomycetes. The steps that most likely need attention when moving to other species combinations are highlighted in the protocol below.
Materials and Reagents
Falcon® tube (50 ml) * 2 (Every similar-sized version will do)
2 ml Eppendorf® tubes (Every similar version will do)
Miracloth® (8 cm x 8 cm) (Millipore, catalog number: 475855-1R)
Arabidopsis thaliana Col-0, wildtype and shrk1 x shrk2 [shrk1 x shrk2 produces multilobed haustoria at higher frequency than the wildtype ( Ried et al., 2019 )]
Hpa Noco2 (Col-0 is susceptible to the Hpa strain Noco2)
Toothpick with pointed ends (see Figure 1)
A white sheet of paper (A5 size, ISO 216)
Several 6.5 cm x 7 cm x 7 cm pots with soil
-
Plastic tray with a transparent lid [38 cm x 24 cm x 18 cm (height includes lid)]
Note: The one we used is not produced anymore. Connex Indoor greenhouse, FLOR79045, is a similar version. Every similar sized box from a gardening supplier with a transparent lid should be suitable.
Soil (Stender GmbH, A210)
Fertilizer (WUXAL® Super 8-8-6)
Counting chamber (Hemocytometer) (Marienfeld, catalog number: 0640110)
-
Spray bottle
Note: A small (20-30 ml volume) spray bottle from the cosmetic store which can produce a fine mist/aerosol is suitable for inoculation. Important! Use the same type for all your experiments and use the same distance and spray time to apply similar spore numbers from experiment to experiment. This may be critical for applying a similar infection pressure in each independent biological repeat.
Ethanol 70% (for sterilization, ethanol of technical grade is sufficient)
Sterile water
Phenol (Roth, catalog number: 0040.1, any brand will do)
Lactic acid (Roth, catalog number: 8460.1, any brand will do)
Trypan blue (Sigma, catalog number: T6146, any brand will do)
35% (v/v) glycerol/water (Roth, catalog number: 7530.4, any brand will do)
0.01% trypan blue solution (see Recipes)
Saturated chloral hydrate solution (2.5 g/ml, see Recipes)
Figure 1. Procedure for growing and infecting the Arabidopsis.
A. Spreading seeds evenly on a white sheet. B. Moisture the tip of the toothpick. C. Pick up one seed at once with a moisture tip. D. Put the seed on the surface of the soil. E. Pick up seeds by your index finger. F. 2-week-old Arabidopsis. G. The 2-week-old Arabidopsis’s leaves are evenly filled with the droplets. Scale bar = 0.8 cm. H. The 7 dpi Hpa infected Arabidopsis. The arrows show the first pair of leaves to collect. Scale bar = 0.8 cm.
Equipment
Scissors with a small and pointy front (Hammacher, HSB 544-09, any version similar to this should be suitable for cutting off infected leaves).
Vortex (Vortex-Genie® 2, one with similar function will do)
Growth chamber (Binder, model: KBWF 720)
Eppendorf ThermoMixer C (or other brand. Any temperature controlled Eppendorf® tube shaker with heating function will do)
-
VHX-6000 digital microscope (Keyence, Osaka, Japan)
Note: The advantage of using the VHX digital microscope is that it can record a large sample area by stitching together individual pictures in x, y and z directions. Any other bright field microscope with a 10x objective will be also suitable but perhaps less comfortable to use for this particular task.
Software
Cell counter (A plugin for the Fiji software, Kurt De Vos, https://imagej.nih.gov/ij/plugins/cell-counter.html)
Procedure
-
Growing of Arabidopsis
-
Prepare Arabidopsis seedlings for the multilobed haustoria observation
-
Stratify all the testing seeds at 4 °C overnight once after you harvest the seeds.
Note: Stratification can synchronize the growing of the plants.
Spread the amount of the seeds that you need onto a white paper (5-9 seeds for one pot) (Figure 1A).
Moisture the tip of the toothpick by dipping it into water (Figure 1B).
-
Distribute 5-9 seeds evenly in one 6.5 cm x 7 x 7 cm pot with the now sticky toothpick tip to pick up one seed each time (Figures 1C-1D).
Notes:
Don’t grow more than 12 plants in this pot size. A crowded pot will lead to an uneven infection because overlapping leaves will cover each other.
Do not cover the seeds with the soil! Put the seeds directly on the surface of the soil.
Grow plants at 22 °C, 16 h light (100 μmol/m2/s)/ 8 h dark, 60% relative humidity for 2 weeks. The plants were watered twice a week with the 1/1000 diluted fertilizer. Figure 1F shows 2-week-old plants grown under the conditions and pot size indicated.
-
-
Prepare Arabidopsis seedlings for Hpa propagation
Stratify all the testing seeds at 4 °C overnight once after you harvest the seeds.
Spread the amount of the seeds that you need onto a white paper (about 50-100 seeds in one pot).
-
Collect the seeds with your index finger and spread them onto the soil evenly like spreading salt on food (Figure 1E).
Note: Do not cover the seeds with the soil! Put the seeds directly on the surface of the soil.
Grow plants at 22 °C, 16 h light (100 μmol/m2/s)/8 h dark, 60% relative humidity for 2 weeks.
-
-
Hpa propagation and preservation
-
Hpa propagation
Sterilize scissors with 70% ethanol.
Clean and decontaminate the plastic tray and lid first with soap (detergent) then with 70% ethanol. Let them dry before further use.
Place the pots which are prepared in Step A2 in the plastic tray and add 250 ml sterilized water in each tray to keep the humidity for the later step.
-
Carefully open the tray of the 7 dpi Hpa infected plants and lift the cover slowly in a defined area for the Hpa infection. Reduce air movement around the plants to a minimum.
Note: If you start the Hpa infection for the very first time in your lab, the Hpa source can be the pots of 7 dpi Arabidopsis or the freezed material (see Step B2) from the provider.
-
Harvest the aerial parts of the infected plants covered with spores (Figure 2A) into a 50 ml Falcon® tube.
Note: The infected Arabidopsis seedlings should be loosely placed in the tube to let the spores easily release to the sterilized water (see Figure 1B in Asai et al., 2015,). Usually, 2-3 full pots of the infected Arabidopsis are used.
Add 15 ml of sterile water in the Falcon® tube and vortex it at the full speed (3,200 rpm) for 10 s.
Filter the suspension using one layer of Miracloth.
Measure the concentration of the conidiospores in the suspension by using the counting chamber (Hemocytometer). Adjust the concentration to 105 spores/ml water. If the spores are not enough, repeat Steps B1d-B1f for the rest of the materials.
Spray the 2-week-old Arabidopsis with the filtered suspension until the leaf surface is evenly filled with the droplets (Figure 1G).
Cover the plastic tray with the transparent lid and seal the openings with transparent tape to maintain high humidity (90%-100%).
-
Incubate the infected plants in a growth chamber at 18 °C, 16 h light (80 μmol/m2/s*, 70 μmol/m2/s under the transparent lid)/8 h dark for 6 or 7 days.
Notes:
*This is the actual measurement from the Binder chamber. The plants grow in the walk in chamber with the light intensity 100 μmol/m2/s, but the incubation has been done in the Binder chamber.
For the propagation and the collection of the fresh conidiospores, Hpa should be maintained on Arabidopsis in a 6- or 7-day-regime. In other words, infect the plants every 6 or 7 days. Usually, Hpa will start to sporulate on 5 dpi. 6 dpi or 7 dpi are the better days for the conidiospores collection.
-
Hpa preservation and regeneration
Follow Steps B1a-B1e to collect the infected plants into a 50 ml Falcon® tube.
-
Preserve the infected plants at -80 °C.
Note: The Hpa conidiospores can be stored in this condition for at least 1 year or longer.
For the regeneration, take out the preserved infected leaves from the -80 °C and put the tube on ice for 10 min.
After 10 min, add 15 ml of sterile water in the Falcon® tube and vortex for 10 s at full speed.
Filter the suspension using one layer of Miracloth.
-
Spray the Arabidopsis (prepared from Step A2) with the filtered suspension until the leaf surface is evenly filled with the droplet (Figure 1G).
Note: Since the viability of the frozen conidiospores will be lower, we don’t adjust (dilute) the concentration for the regeneration.
-
-
Hpa inoculation and incubation
-
Inoculation
Randomly place different genotypes of Arabidopsis pots prepared from Step A1 in the tray.
Infect the plants as in Step B1.
-
Incubation
Incubate the infected plants in a growth chamber at 18 °C, 16 h light (80 μmol/m2/s, 70 μmol/m2/s under the transparent lid)/ 8 h dark until the collection day.
Note: Under these conditions, in the Arabidopsis thaliana Col-0 x Hpa Noco2 interaction, the multilobed haustoria start to accumulate on 5 dpi (Figure 3) and reach to a higher proportion at later infection stages (7 dpi-8 dpi) in both wild type and the shrk1 x shrk2 mutant.
-
-
Staining of plant tissues for visualization of the pathogen
-
Harvest one of the infected first pair of true leaves from each seedling (5-8 biological repeats for each genotype= 5-8 plants) (Figure 1H).
Note: For other oomycete-plant pathosystems, the choice of leaf may vary. If nothing else matters, pick the youngest infected leaves as they are typically the smallest and easiest to stain.
-
Incubate them in the trypan blue solution which cover the leaves in 2 ml Eppendorf® tubes at 95 °C for 3-10 min or until the tissues completely soaked up the solution and incubate them at room temperature overnight.
Notes:
To avoid the splitting of the trypan blue while cooking the samples, don’t put trypan blue solution excess than 1 ml in the 2 ml Eppendorf®.
Do not heat them longer than 30 min, the tissues will become too soft for further steps. However, the room temperature overnight process can be extended if the tissues are not stained enough. Roll the Eppendorf tube a bit to see whether the tissue is well soaked by the trypan blue. The ready tissues should be dark blue (Figure 2B).
For the other oomycete pathosystems, heat and incubate the samples until the tissues completely soaked up the solution. The required incubation time may increase with leaf and cell wall thickness.
-
Replace the solution with the saturated chloral hydrate solution and leave overnight until the tissues look clear (Figure 2C).
Note: If the tissues are not yet clear, replace the chloral hydrate supernatant with a fresh aliquot and incubate until the tissues become clear.
-
Replace the chloral hydrate with 35% glycerol.
Note: The samples can be stored in 35% glycerol for long time (at least 3 months).
Place the tissues on microscopy slides with 35% glycerol for further observation under the Keyence VHX digital microscope. Use a cover slip to prevent the sample from drying out.
-
-
An introduction of the Keyence VHX microscope and Fiji plugin of ImageJ for the quantification of structures of interest
-
The using of the Keyence VHX digital microscope
-
Turn on the microscope and follow the instruction of the software to position the sample in focus.
Note: For the example showing below, we used 500x objective lens to visualize the samples.
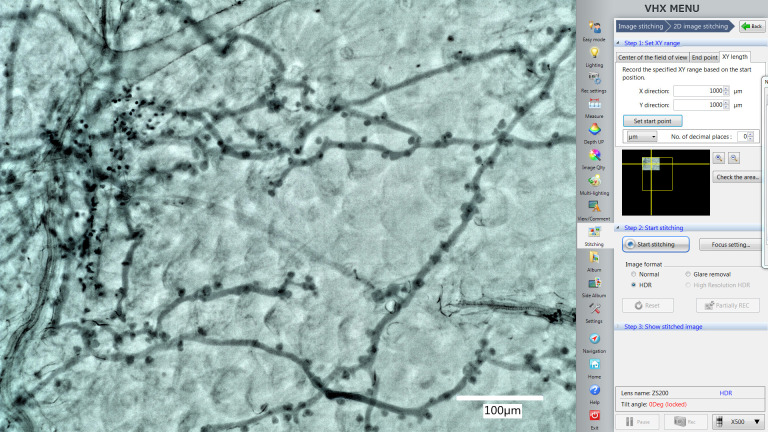
Use the “Stitching” function in the software to acquire pictures that contain larger areas.
-
Use the “XY length” function in the “Stitching” window to set the area you want to include them in one picture (Figure 4, X direction 1,000 µm, Y direction 1,000 µm). The yellow square will also show you the area microscope will record this time.
Note: X direction 1,000 µm and Y direction 1,000 µm is a suggested area size that works well for Arabidopsis leaves at the indicated dpi. You can change the area according to the biological system requirements, but always maintain one consistent size within the same experiment.
-
Take pictures from 5 different and not overlapping areas from one leaf.
Note: Take pictures continually by using the stitching function. It will show you the area you’ve just imaged. This guide should help to avoid overlaps between samples (Figure 5).
-
-
Quantification of multilobed and round-shaped haustoria by the Fiji plugin: cell counter
Note: For a guide how to distinguish round-shaped (“normal”) and multi-lobed haustoria, see: Ried et al., 2019, Figure 2 or the figures taken by the VHX digital microscope in this protocol: Figure 3.
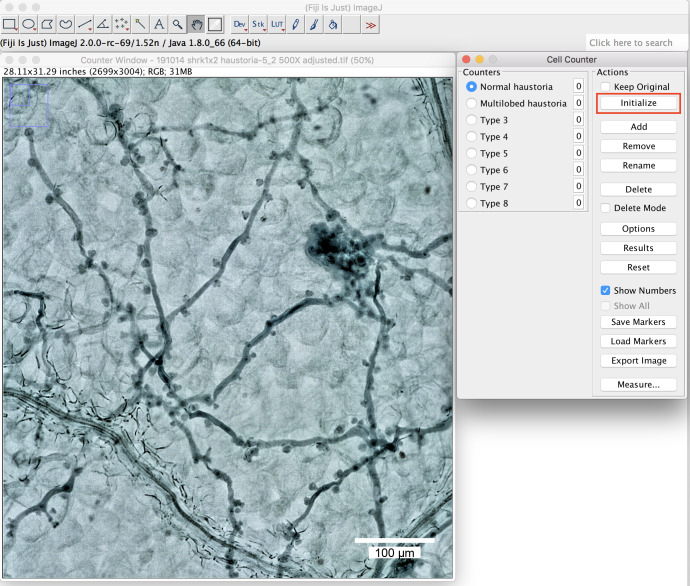
Use the “cell counter” plugin in the Fiji package to count and record the haustoria shape.
Open the picture in Fiji, click on the window of the picture and then start the “cell counter” plugin. Press “Initialize” to activate the picture you want to quantify (Figure 6).
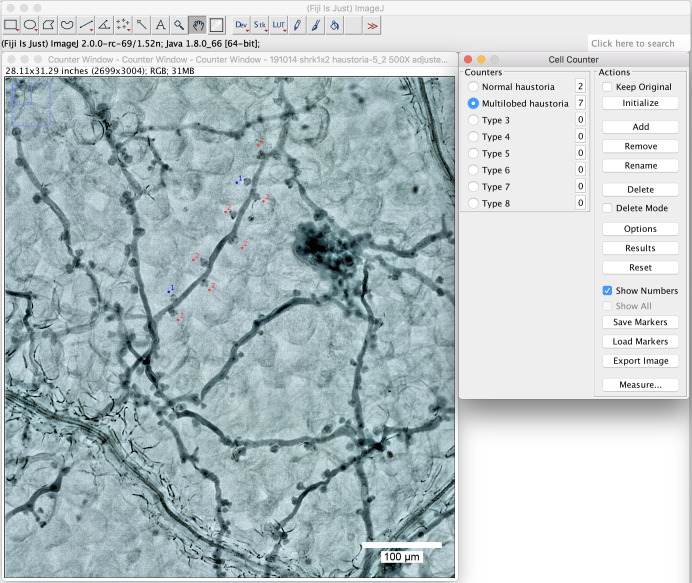
Define the name at the “Counters” (Figure 6 above, round-shaped (normal) haustoria; multilobed haustoria).
-
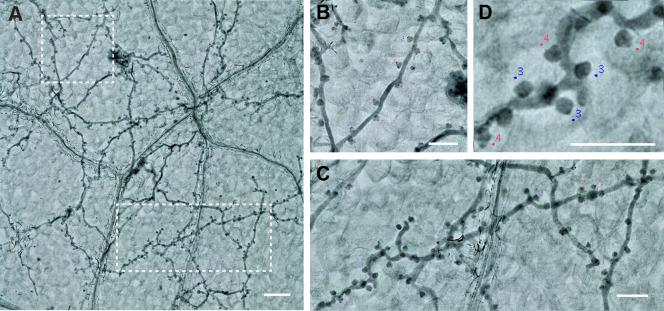
Label them with a simple click beside the haustoria you want to label. Different types will then be labeled in different colors (Figure 7, normal haustoria in blue, multilobed haustoria in red).
Note: Label only the haustoria which are in focus. Ignore the ones for which the shape is difficult to judge.
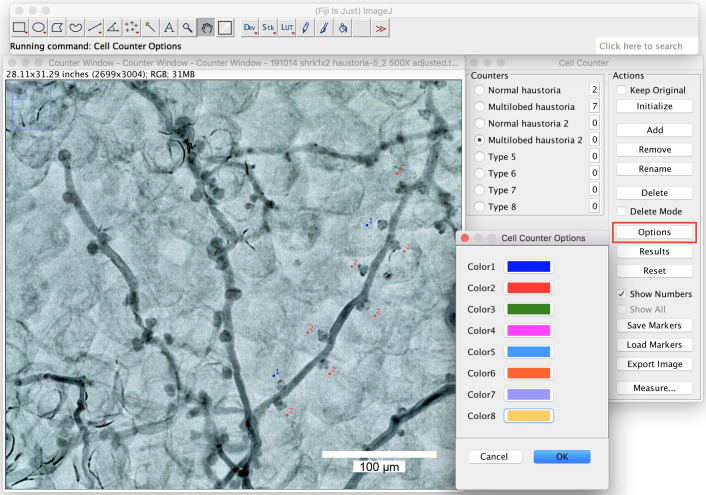
You can change the labeling colors by clicking on the “options” (Figure 8).
Pick up 2 or more branches from one picture to count the proportion of the multilobed haustoria (Figure 9). At least 10 branches in total should be counted in one biological repeat (one leaf).
You can save the labels with the “Save Markers” function.
-
Figure 2. Hpa infected Arabidopsis and the trypan blue stained leaf.
A. The non-infected mock leaf (left) and the 7 dpi Hpa infected leaf (right). Scale bar = 2 mm. B. The trypan blue-stained leaf before the de-staining process. Scale bar = 1 mm. C. The de-stained trypan blue-treated leaf. The hyphae can be clearly seen under the microscope or by the eyes. Scale bar = 1 mm.
Figure 3. The Hpa haustoria in the Col-0 and the shrk1 x shrk2 leaf.
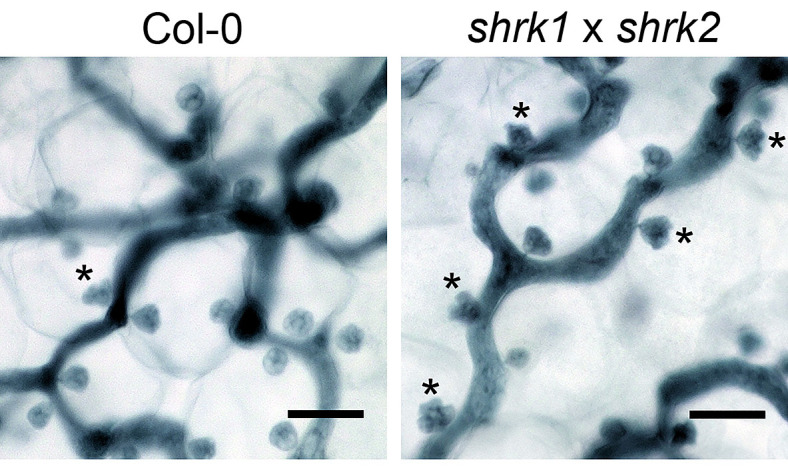
The pictures were taken from the 5 dpi leaves by the 1,000x objective lens with the VHX digital microscope. The asterisks indicate the multilobed haustoria. Scale bars = 25 µm.
Figure 4. The Stitching function in the VHX digital microscope software.
The yellow square shows you the area microscope will record this time. The picture in the yellow square shows the position of the area which is displayed by the live imaging window now.
Figure 5. The Stitching function helps to avoid overlaps between samples.
The yellow square shows us the area just taken. This area should be excluded from additional stitchings from the same leaf to avoid overlapping samples.
Figure 6. The plugin “cell counter” in the Fiji package.
Initialization of the picture. Scale bar =100 μm.
Figure 7. Use the plugin “cell counter” to label the haustoria.
Label the haustoria by a simple click beside the haustoria you want to label. Scale bar = 100 μm.
Figure 8. Change the labeling colors.
Big ranges of the labeling colors can be chosen from the option window by right click on the colors. Scale bar = 100 μm.
Figure 9. The labeling of the Haustoria.
A. Two branches from one picture. Scale bar = 100 µm. B and C. The magnification of the two branches. Scale bar = 50 µm. D. Haustoria labeled with different colors, green: normal haustoria, magenta: multilobed haustoria. Scale bar = 50 µm.
Recipes
-
0.01% trypan blue solution
10 ml lactic acid*
10 ml glycerol
10 g phenol*
10 ml sterile water
10 mg trypan blue
Note: Handle the materials labeled with asterisk cautiously in the chemical hood. Trypan blue is light sensitive. The solution should be stored in a brown bottle, it can be reused at least two times.
-
Chloral hydrate solution (2.5 g/ml)
Add 40 ml water to 100 g chloral hydrate in a 100 ml bottle
Note: 100 g chloral hydrate will already occupy almost the entire space of a 100 ml bottle, therefore; don’t put more than this amount in a 100 ml bottle.
Acknowledgments
The initial establishment of the Hpa x Arabidopsis pathosystem in the author’s laboratory was largely based on Hpa genotypes and advise given by the team of Jane Parker, while still working at The Sainsbury Laboratory in Norwich. The Steps B1 and D described in this protocol are similar to the one described by Asai et al. (2015). Procedures A-D were used as described in Ried et al. (2019). MP acknowledges funding by the Deutsche Forschungsgemeinschaft (DFG) and the European Research Council (ERC).
Competing interests
The authors have no competing interests to declare.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Asai S., Shirasu K. and Jones J. D. G.(2015). Hyaloperonospora arabidopsidis(Downy Mildew) Infection Assay in Arabidopsis . Bio-protocol 5(20): e1627. [Google Scholar]
- 2. Caillaud M. C., Wirthmueller L., Sklenar J., Findlay K., Piquerez S. J., Jones A. M., Robatzek S., Jones J. D. and Faulkner C.(2014). The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog 10(10): e1004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Callan B. and Carris L.(2004). Biodiversity of Fungi: Inventory and Monitoring Methods. Elsevier Inc. pp: 113-114 [Google Scholar]
- 4. Judelson H. S. and Ah-Fong A. M. V.(2019). Exchanges at the Plant-Oomycete Interface That Influence Disease. Plant Physiol 179(4): 1198-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Agrios N. G.(2005). Plant Pathology. 5th edition. Elsevier Inc. pp. 427-433. [Google Scholar]
- 6. Ried M. K., Banhara A. and Hwu F. Y., Binder A., Gust A. A., Höfle C., Hückelhoven R., Nürnberger T., Parniske M.(2019). A set of Arabidopsis genes involved in the accommodation of the downy mildew pathogen Hyaloperonospora arabidopsidis . PLoS Pathog 15(7): e1007747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P. and Cardona A.(2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7): 676-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang W., Wen Y., Berkey R. and Xiao S.(2009). Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal Haustorium renders broad-spectrum resistance to powdery mildew . Plant Cell 21(9): 2898-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]