Figure 2.
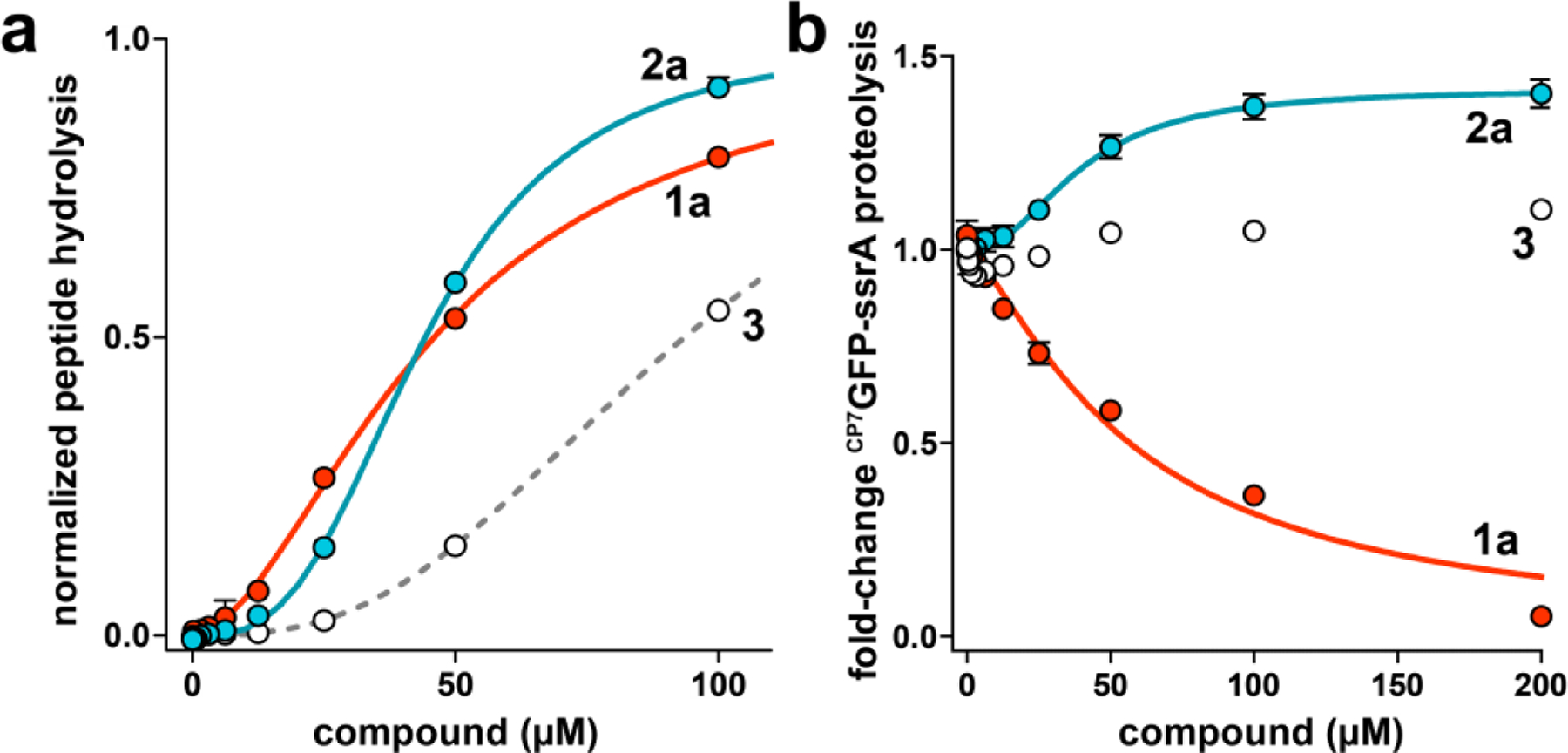
ADEP and fragments stimulate ClpP1P2 peptidase activity, but have different effects on ClpXP1P2 proteolysis activity. (a) Hydrolysis of a fluorogenic decapeptide (Abz-KASPVSLGYNO2D; 15 μM) by 0.5 μM ClpP1P2 was stimulated by ADEP 1a (red circles), fragment 2a (blue circles), and fragment 3 (white circles). Data were fit to a Hill equation. 1a stimulated peptidase activity with EC50 = 38 ± 2 μM, Hill constant (n) = 1.9 ± 0.07 (solid red line); 2a with EC50 = 46 ± 0.4 μM, n = 3.3 ± 0.2 (solid blue line); and 3 with EC50 = 94 ± 4 μM, n = 2.7 ± 0.1 (dashed gray line). (b) 1a inhibited proteolysis of 10 μM CP7GFP-ssrA by 1 μM ClpXP1P2 with IC50 = 59 ± 9 μM, n = 1.3 ± 0.2 μM. 2a stimulated with EC50 = 40 ± 5 μM, n = 2.1 ± 0.4, to a maximum activity 1.4-fold above the basal proteolysis rate. As fragment 3 had little effect on proteolysis over the tested concentration range, data were not fit to a binding equation. Values in all panels are averages of three biological replicates (N = 3) ± 1 standard deviation (SD).
