Figure 3.
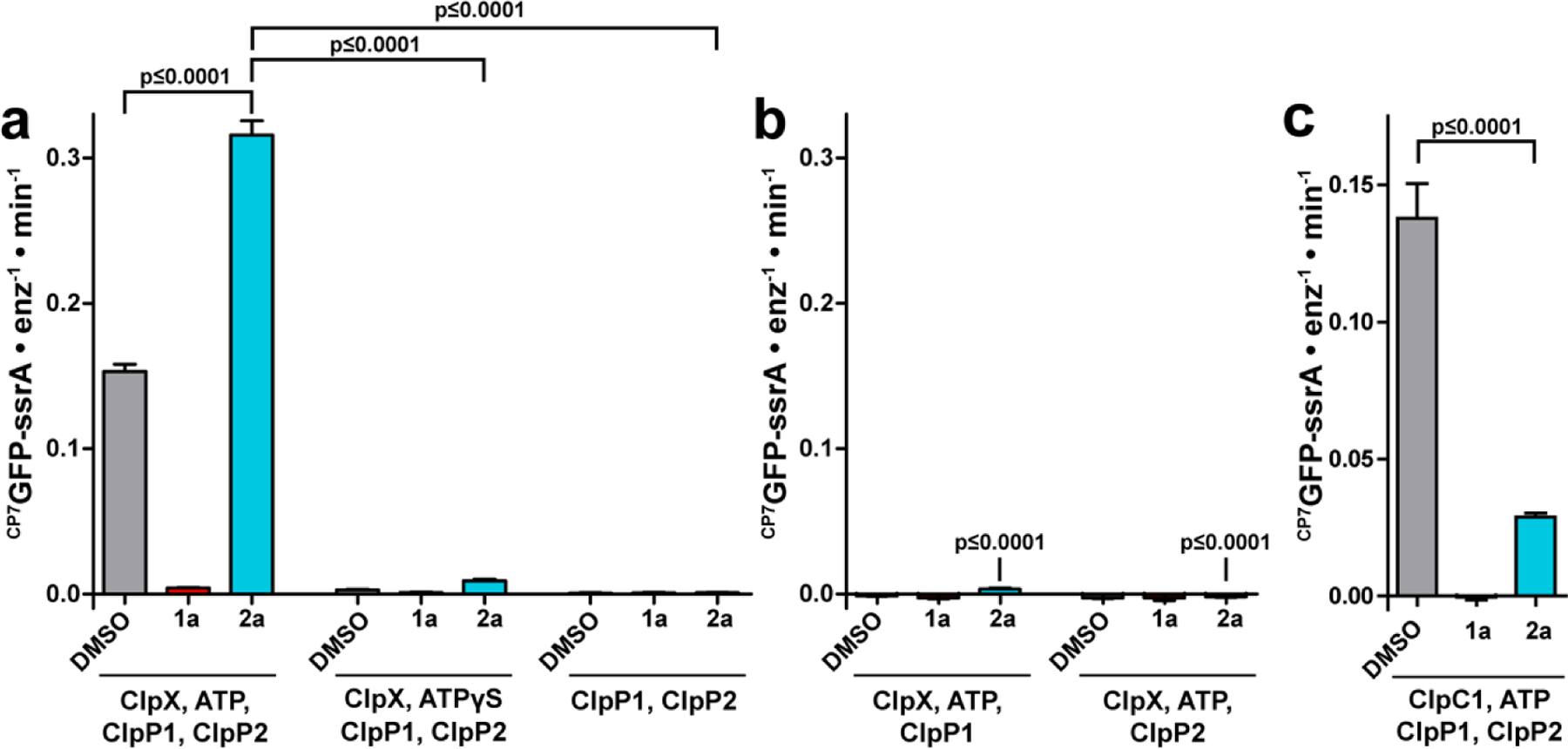
Fragment 2a stimulates canonical ATP-dependent proteolysis by ClpXP1P2. Proteolysis of 10 μM of the indicated substrate was assayed in the presence of DMSO vehicle, 200 μM 1a, or 200 μM 2a. (a) Proteolysis and stimulation by 2a requires ClpX and ATP. Degradation of CP7GFP-ssrA was observed in the presence of 2 μM ClpP1P2, 1 μM ClpX, and 2.5 mM ATP together with an ATP regeneration system. Proteolysis was inhibited by 1a and stimulated by 2a. Weak proteolysis occurred in the presence of 2.5 mM ATPγS, and this activity was stimulated by 2a. No activity was detected in the presence of ClpP1P2 alone. (b) Proteolytic stimulation by 2a requires both ClpP1 and ClpP2. Degradation of CP7GFP-ssrA was not observed when 1 μM ClpX, 2.5 mM ATP and an ATP regeneration system were combined with ClpP1 or ClpP2 individually. (c) Degradation of CP7GFP-ssrA by 1 μM ClpC1, 2 μM ClpP1P2, and an ATP regeneration system was partially inhibited by 2a. Values in all panels are averages of three technical replicates (N = 3) ± 1 SD. p-values were calculated by unpaired two-tailed Student’s t-test. The p-values in panel b are compared to the leftmost condition in panel a.
