Figure 4.
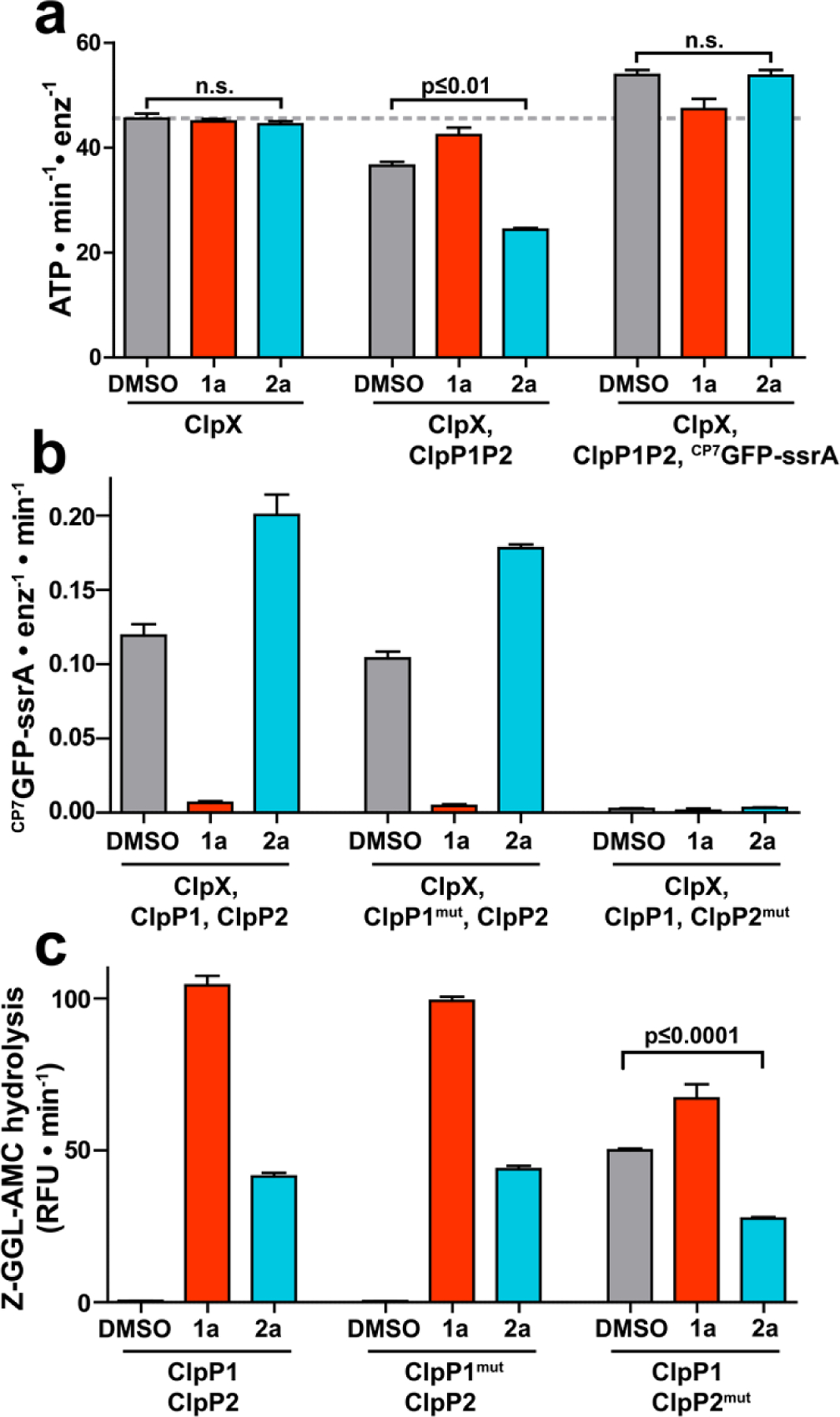
Fragment 2a binds to a second site on ClpP1P2. (a) The ATPase activity of 1 μM ClpX (left group), 1 μM ClpX with 2 μM ClpP1P2 (center group), or 1 μM ClpX with both 2 μM ClpP1P2 and 10 µM CP7GFP-ssrA (right group) was measured in the presence of DMSO vehicle, 200 μM 1a, or 200 μM 2a. Neither 1a nor 2a altered the basal ClpX ATPase activity. 1a, but not 2a, blocked ATPase suppression by ClpP1P2 and ATPase stimulation by substrate. (b) Proteolysis of CP7GFP-ssrA by 1 µM ClpX and 2 µM ClpP1P2 incorporating LGF-pocket substitutions in the indicated subunits was measured in the presence of DMSO control, 200 µM compound. 1a inhibited and 2a stimulated proteolysis by wild-type enzyme and enzyme incorporating ClpP1mut. No proteolysis was observed by enzyme incorporated ClpP2mut, which weakens ClpX binding. (c) Hydrolysis of a fluorogenic tripeptide (Z-Gly-Gly-Leu-AMC; 50 μM) by 0.5 μM ClpP1P2 incorporating wild-type subunits or LGF-pocket substitutions was assayed in the presence of 200 μM 1a or 2a. Peptidase incorporating ClpP2mut exhibited peptidase activity in the absence of compound, which was stimulated by 1a and suppressed by 2a. p-values were calculated by unpaired two-tailed Student’s t-test; n.s. = not significant. Values in all panels are averages of three technical replicates (N = 3) ± 1 SD.
