Figure 5.
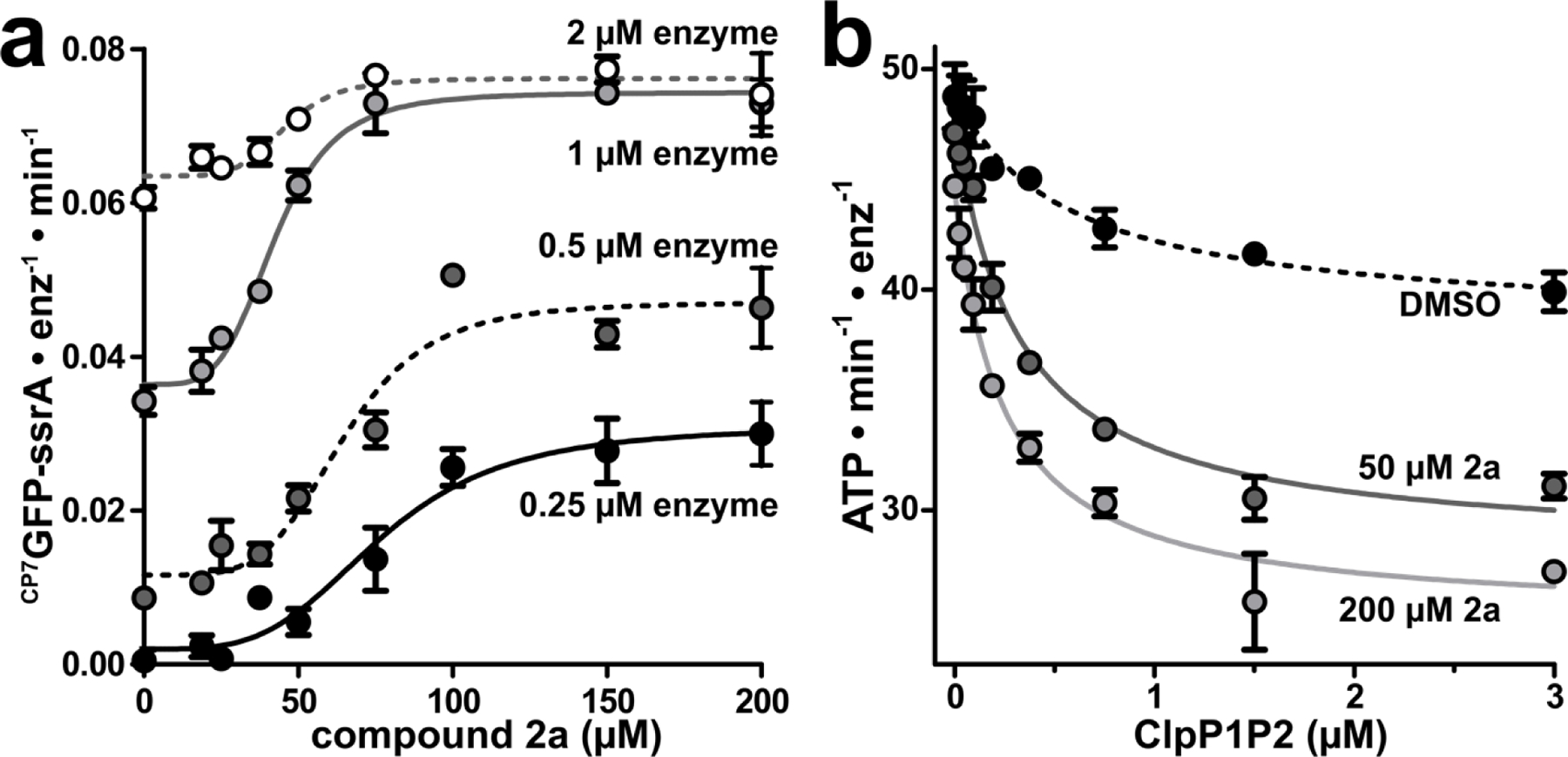
Fragment 2a stabilizes ClpXP1P2 assembly. (a) CP7GFP-ssrA proteolysis was measured as a function of 2a concentration for four concentrations of ClpXP1P2. Data were fit to a Hill equation. 0.25 μM ClpXP1P2 stimulated activity ~16-fold with EC50 = 77 ± 6 μM, n = 3.7 ± 1; 0.5 μM enzyme stimulated ~4-fold with EC50 = 65 ± 4 μM, n = 4.7 ± 1; 1 μM enzyme stimulated 2-fold with EC50 = 42 ± 2 μM, n = 4.4 ± 0.7; and 2 μM stimulated 1.2-fold with EC50 = 46 ± 4 μM, n = 5.5 ± 3. (b) The ATPase activity of 0.25 μM ClpX was measured as a function of ClpP1P2 concentration in the presence of DMSO mock (black circles), 50 μM 2a (dark gray circles), or 200 μM 2a (light gray circles). Data were fit to a non-cooperative binding model. ClpP1P2 suppressed ClpX ATP hydrolysis to ~80% of basal activity in the presence of DMSO with a Kapp of 0.57 ± 0.2 μM; to ~60% in the presence of 50 μM 2a with Kapp 0.31 ± 0.04 μM; and to ~55% in the presence of 200 µM 2a with Kapp 0.24 ± 0.03 μM. Fragments with KD-ClpP1 << KD-ClpP2 may stimulate proteolysis at concentrations below KD-ClpP2. p-values were calculated by unpaired two-tailed Student’s t-test; n.s. = not significant. Values in all panels are averages of three technical replicates (N = 3) ± 1 SD.
