Figure 6.
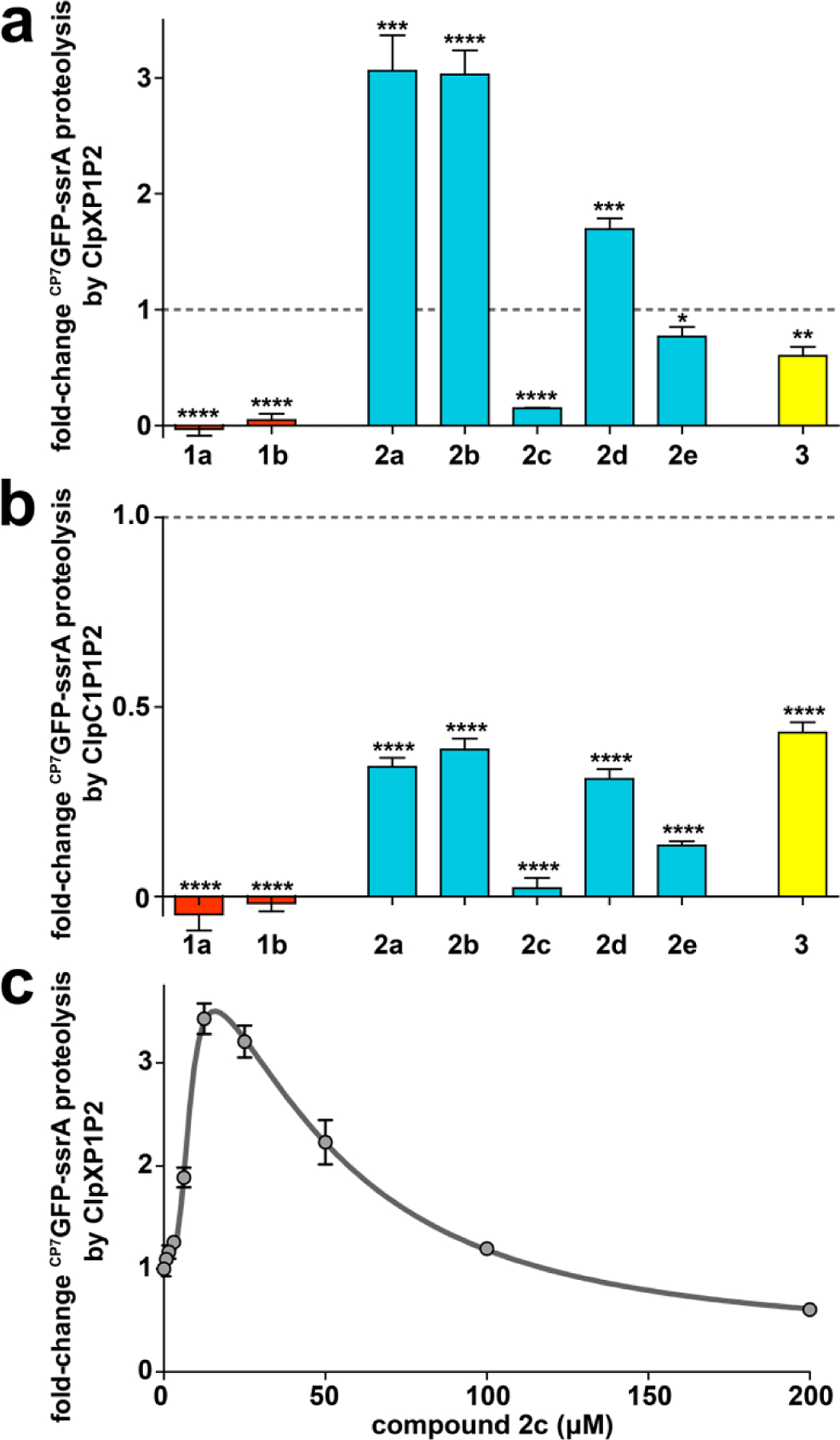
ADEP fragments may stimulate or inhibit proteolysis. (a) Distinct fragments have different effects on proteolysis by ClpXP1P2. Proteolysis of 10 μM CP7GFP-ssrA by 0.75 μM ClpX, 2 μM ClpP1P2, and an ATP regeneration system was assayed in the presence of 200 μM of the indicated compound (except for 1b, which was assayed at 50 μM). Activities were normalized to that of a DMSO vehicle control (dashed horizontal line). (b) All fragments inhibit proteolysis by ClpC1P1P2 to varying extents. Proteolysis of CP7GFP-ssrA (10 μM) by 0.75 μM ClpC1, 2 μM ClpP1P2, and an ATP regeneration system was assayed as in panel a. (c) Proteolysis of CP7GFP-ssrA (10 μM) by 1 μM ClpX, 2 μM ClpP1P2, 2.5 mM ATP and a regeneration system was measured as a function of 2c concentration. Data were fit to a double Hill equation (see Supporting Information). 2c stimulated proteolysis at low concentrations with EC50–1 = 7.8 ± 0.8 μM, n1 = 3.8 ± 1, and inhibited proteolysis at high concentrations with EC50–2 = 50 ± 10 μM, n2 = 1.8 ± 0.6. Values in all panels are averages of three technical replicates (N = 3) ± 1 SD. p-values were calculated by unpaired two-tailed Student’s t-test: * p≤0.05; ** p≤0.01; *** p≤0.001; **** p≤0.0001.
