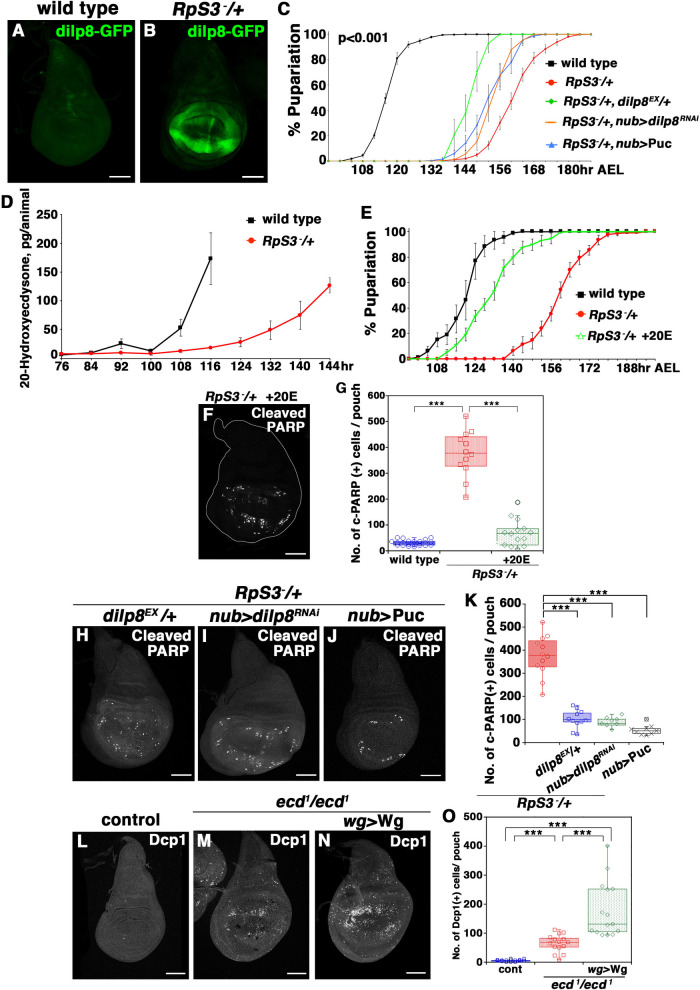
Fig 4. JNK-Dilp8-mediated developmental delay is required for massive cell-turnover.
(A and B) The expression of dilp8 in wing discs of dilp8::eGFP/+ (A) or RpS3/+, Dilp8::eGFP/+ (B) flies were visualized with GFP (green). Scale bar, 100 μm. (C) Pupariation curves for wild-type (n = 390), RpS3/+ (n = 666), RpS3/+, dilp8EX/+ (n = 46), RpS3/+, nub>dilp8RNAi (n = 215), or RpS3/+, nub>Puc (n = 302) flies. p>0.001, comparing RpS3/+ to the other flies (log-rank analysis). Error bars, SEM. (D) 20-Hydroxyecdysteroid titer was measured in wild-type or RpS3/+ larvae. (E) Pupariation curves of wild-type or RpS3/+ larvae that were fed food containing 0.75 mg/ml 20E or control. Error bars, SEM. (F) Dying cells were visualized by anti-cleaved PARP staining (white) in the wing discs of RpS3/+ larvae that were fed food containing 0.75 mg/ml 20E. Scale bar, 100 μm. (G) Boxplot with dots representing cleaved-PARP-positive dying cells per pouch in genotypes shown in (Fig 3A) (n = 22), (Fig 3B) (n = 12), and (F) (n = 14). Error bars, SEM; ***, p<0.001; non-parametric Mann-Whitney U-test. (H-I) Dying cells were detected by anti-cleaved PARP staining in the wing discs of RpS3/+, dilp8EX/+ (H), RpS3/+, nub-Gal4, UAS- dilp8-RNAi (I), or RpS3/+, nub-Gal4, UAS-Puc (J) flies expressing CD8-PARP-Venus. Scale bar, 100 μm. (K) Boxplot with dots representing cleaved-PARP-positive dying cells per pouch in genotypes shown in (Fig 3B) (n = 12), (H) (n = 12), (I) (n = 9), and (J) (n = 9). Error bars, SEM; ***, p<0.001; non-parametric Mann-Whitney U-test. (L-N) Dying cells were visualized by anti-cleaved Dcp-1 staining in the wing discs (white). ecd1 is a temperature-sensitive ecd mutant allele that blocks biosynthesis of the active-form of the hormone 20-Hydroxyecdysone at 29°C. Non-heat-shock treated flies was grown at 18°C (L). For heat-shock treatment, ecd1/ecd1 (M), or ecd1/ecd1, wg-Gal4, UAS-Wg (N) fly culture was transferred to 29°C for 48 hours during the 3rd instar larval stage. (O) Boxplot with dots representing cleaved-Dcp-1-positive dying cells per pouch in genotypes shown in (L) (n = 14), (M) (n = 15) and (N) (n = 15). Error bars, SEM; ***, p<0.001; non-parametric Mann-Whitney U-test. Scale bar, 100 μm. Genotypes are as follows: dilp8::eGFP/+ (A), RpS3Plac92/dilp8::eGFP (B), nub-Gal4/+; UAS-CD8-PARP-Venus/+ (black), nub-Gal4/+; UAS-CD8-PARP-Venus, RpS3Plac92/+ (red), nub-Gal4/+; UAS-CD8-PARP-Venus, RpS3Plac92/dilp8EX (green), nub-Gal4/UAS-dilp8-RNAi; UAS-CD8-PARP-Venus, RpS3Plac92/+ (orange), nub-Gal4/+; UAS-CD8-PARP-Venus, RpS3Plac92/UAS-Puc (blue) (C), w1118 (D: wild-type), FRT82B, Ubi-GFP, RpS3Plac92/+ (D; RpS3/+), nub-Gal4/+; UAS-CD8-PARP-Venus/+ (E: wild-type), nub-Gal4/+; UAS-CD8-PARP-Venus, RpS3Plac92/+ (E: RpS3/+, F), nub-Gal4/+; UAS-CD8-PARP-Venus, RpS3Plac92/dilp8EX (H), nub-Gal4/+; UAS-CD8-PARP-Venus, RpS3Plac92/UAS-dilp8-RNAi (I), nub-Gal4/+; UAS-CD8-PARP-Venus, RpS3Plac92/UAS-Puc (J), ecd1/ecd1 (L, M), and wg-Gal4/UAS-Wg; ecd1/ecd1 (N).