Figure 2.
RBM functional constraints compared to RBM natural diversity
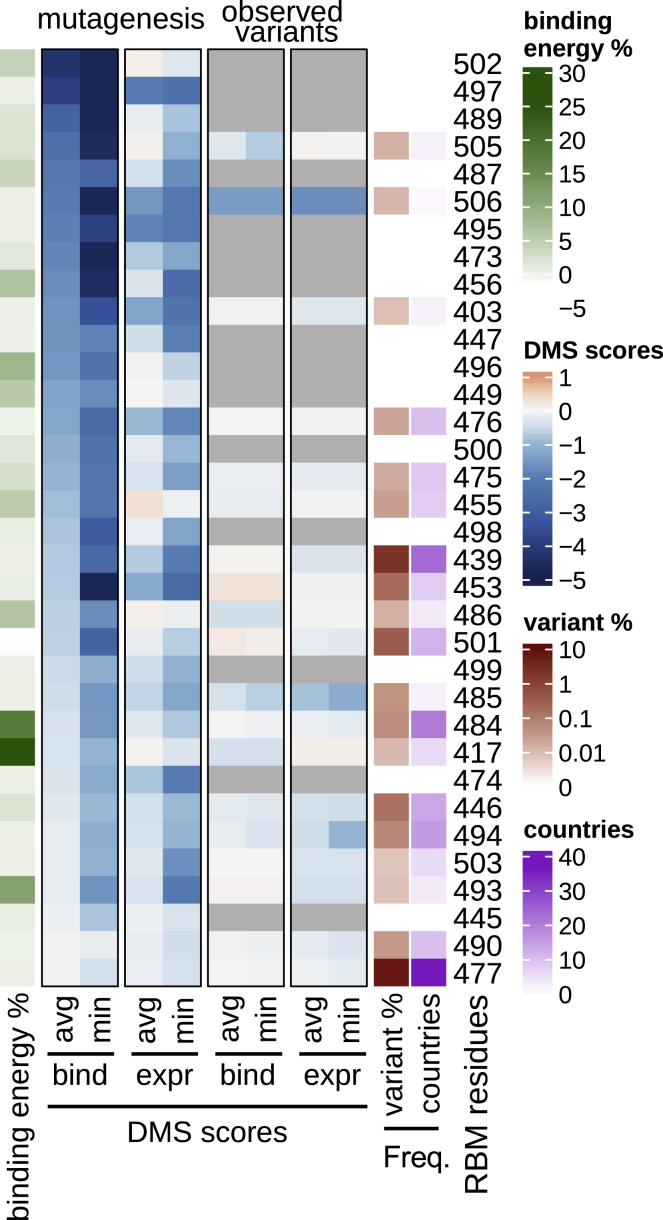
Each residue in the RBM is annotated by several metrics, depicted as a heatmap. DMS scores: outlined in black boxes (center) are summaries of hACE2 binding and RBD expression deep mutational scanning (DMS) experimental results (Starr et al., 2020b). DMS score is the binding or expression fold change of a variant over WT on a log10 scale (red indicating improvement and blue indicating loss as compared to WT). In the “mutagenesis” columns, DMS results are given for each residue as either the minimum (most disruptive variant) or the average score across all possible variants of a residue, except for the reference residue and the stop codon. In the “observed variants” columns, minimum and average scores are computed only across variants that have been observed in GISAID (same set of sequences as used for Figure 1). When no natural variants have been observed, cells are gray. Data were sorted on the leftmost DMS column. Frequency: each RBM position is annotated with the frequency of non-reference amino acids in deposited sequences (darker red indicating higher frequency; at least 1 supporting sequence per 25,000 deposited sequences is required to call a variant). The number of countries in which variants have been observed is also annotated (darker purple indicating more countries). Binding energy: a re-refined SARS-CoV-2 RBD:hACE2 complex X-ray structure (PDB: 6M0J) was used to determine the approximate, decomposed binding free energy associated with each RBM residue. Results for each RBM residue are expressed as a percentage of the total binding interface interaction energy (darker green indicating stronger contribution to the binding energy).
See also Figures S1 and S2.