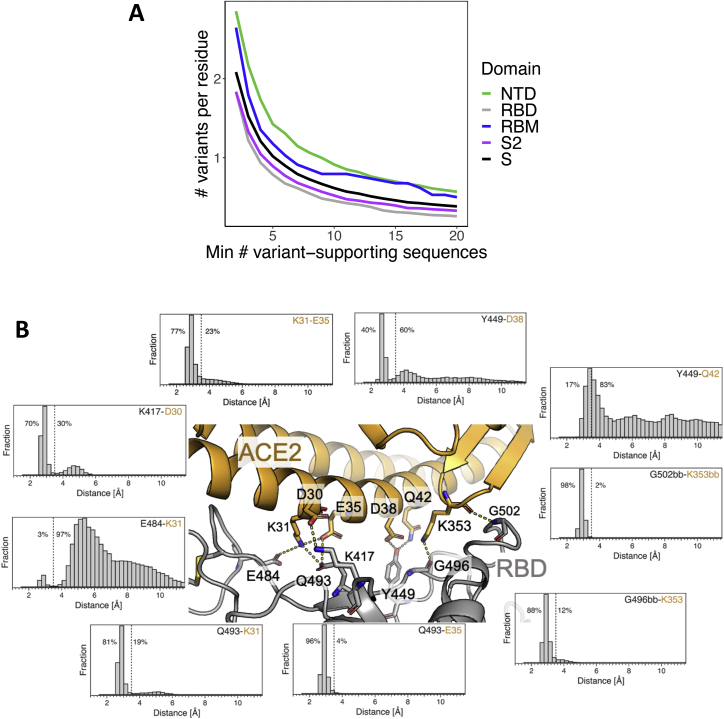
Figure S1.
High RBM variability in deposited SARS-CoV-2 sequences is consistent with a dynamic RBD:hACE2 binding interface, related to Figures 1 and 2
(A) Number of observed variants in four S domains (or full mature S protein) normalized by the total number of residues in each domain, where the number of observed isolates required to call a variant is varied along the x axis.
(B) Distributions of distances observed for RBD (gray):hACE2 (gold) residue pairs: K417-D30, E484-K31, Q493-K31, Q493-E35, G496bb-K353, G502bb-K353bb, Y449-Q42, Y449-D38, K31-E35 (bb = backbone interaction). RBD:ACE2 residue pairs were chosen based on RBM residues with high binding energies as determined by the binding energy % column (green) in Figure 2. Distances were computed every 2.5 ns from 118.7 μs of molecular dynamics simulation data. Dashed lines indicate a distance of 3.5 Å and the percentage of distances below and above 3.5 Å are annotated to the left and right of the lines, respectively.