Abstract
Background
The present COVID-19 pandemic has prompted worldwide repurposing of drugs. The aim of the present work was to develop and validate a two-dimensional isotope-dilution liquid chromatrography tandem mass spectrometry (ID-LC–MS/MS) method for accurate quantification of remdesivir and its active metabolite GS-441524, chloroquine, hydroxychloroquine, lopinavir, ritonavir, favipiravir and azithromycin in serum; drugs that have gained attention for repurposing in the treatment of COVID-19.
Methods
Following protein precipitation, samples were separated with a two-dimensional ultra-high performance liquid chromatography (2D-UHPLC) setup, consisting of an online solid phase extraction (SPE) coupled to an analytical column. For quantification, stable isotope-labelled analogues were used as internal standards for all analytes. The method was validated on the basis of the European Medicines Agency bioanalytical method validation protocol.
Results
Detuning of lopinavir and ritonavir allowed simultaneous quantification of all analytes with different concentration ranges and sensitivity with a uniform injection volume of 5 μL. The method provided robust validation results with inaccuracy and imprecision values of ≤ 9.59 % and ≤ 11.1 % for all quality controls.
Conclusion
The presented method is suitable for accurate and simultaneous quantification of remdesivir, its metabolite GS-441525, chloroquine, hydroxychloroquine, lopinavir, ritonavir, favipiravir and azithromycin in human serum. The quantitative assay may be an efficient tool for the therapeutic drug monitoring of these potential drug candidates in COVID-19 patients in order to increase treatment efficacy and safety.
Keywords: Antiviral therapy, Therapeutic drug monitoring, Isotope dilution liquid chromatography tandem mass spectrometry (ID-LC–MS/MS)
1. Introduction
The present coronavirus disease 2019 (COVID-19) pandemic is a global challenge for health care systems. Most people infected with SARS-CoV-2, the virus that causes COVID-19, develop mild to moderate symptoms and recover on their own. Some patients with COVID-19, however, develop severe symptoms and require hospitalization up to the point of intensive care therapy.
Corticosteroid therapy with dexamethasone proved to have some beneficial effect on severely ill COVID-19 patients [1]. So far, no other drugs have been shown to be effective for specific treatment of COVID-19 and optimized supportive care remains the mainstay of therapy. In view of the high urgency, considerable attention has been focused on the repurposing of drugs with in vitro antiviral activity against SARS-CoV-2. Several substances are considered as potential drug candidates and therefore subject to ongoing clinical research [2].
Of particular note is investigational COVID-19 drug remdesivir, a promising broad-spectrum antiviral agent originally developed to treat hepatitis C, where special approvals have been granted for the treatment of severe illness from COVID-19 [3]. Other potential drug candidates are the antiviral substance favipiravir that is used to treat influenza [4], and fixed-dose combination medication of the antiviral lopinavir and its pharmacological booster ritonavir (potent CYP3A4 inhibitor) for the prevention and treatment of HIV [5]. Azithromycin, a broad-spectrum antibiotic with antiviral and immunomodulatory properties that is primarily eliminated as an unchanged drug, also represents an interesting candidate in the search for COVID-19 drug therapy [6]. Chloroquine und hydroxychloroquine that are used for the treatment of malaria and autoimmune disorders such as systemic lupus erythematosus were temporarily withdrawn for treatment of COVID-19 given that large-scale trials have failed to show any survival benefit [7]. However, due to the usage of unreliable data in some studies the WHO resumed the hydroxychloroquine arm of COVID-19 Solidarity trial.
Typically, the dosage of repurposed COVID-19 therapeutics is derived from in vitro generated half maximum effective concentration (EC50) values for SARS-CoV-2 and pharmacology-based pharmacokinetic models previously developed in other diseases and clinical conditions. However, corresponding dosage regimens will not necessarily translate into adequate drug exposure in critically ill COVID-19 patients due to pathophysiological alterations [8]. It is conceivable that dosage regimens of repurposed drugs may lead to subtherapeutic or toxic concentrations without clear clinical benefit. Accordingly, there is an urgent need to generate high-quality pharmacokinetic data for drug repurposing against COVID-19 [9]. In addition, polytherapy can result in unpredictable drug levels due to relevant drug-drug interactions. Consideration should therefore be given to an individualized optimal dosing strategy, where therapeutic drug monitoring (TDM) can further contribute to establish efficacy and safety.
Different quantitative methods have been described in the literature for analysis of individual components or part of the above-mentioned repurposed COVID-19 drugs. Recently, an UHPLC tandem mass spectrometry (MS/MS) assay was published for the simultaneous quantification of the prodrug remdesivir and its active metabolite GS-441524 [10] that exhibits a considerably longer elimination half-life (t½) of approximately 24 h [11]. Several assays are described for the quantification of repurposed COVID19 drugs, such as HPLC with a diode-array detector (DAD) and LC–MS/MS for chloroquine [12,13], LC–MS/MS for hydroxychloroquine [14,15], HPLC coupled to UV detection and LC–MS/MS for lopinavir, ritonavir [16,17], favipiravir [18,19] and LC–MS/MS for azithromycin [20,21]. Especially, isotope dilution liquid chromatography tandem mass spectrometry (ID-LC–MS/MS) with application of stable isotope labelled analogues as internal standard is considered the gold standard in TDM [22]. However, to our knowledge no multi-analyte isotope dilution ID-LC–MS/MS method provides simultaneous quantification of remdesivir, and its metabolite GS-441524, chloroquine, hydroxychloroquine, lopinavir, ritonavir, favipiravir and azithromycin in a single analytic run with two different separation stages.
Two-dimensional liquid chromatography (2D-LC), where the eluate of a first column (the first dimension) is transferred to a second column (the second dimension), is becoming more widely adopted in the analysis of pharmaceutical compounds, especially when dealing with complex samples [23]. Therefore, the main objective of the present study was to establish a quantitative multi-analyte ID-LC–MS/MS method for these pharmaceuticals in human serum. To allow a high sample throughput with minimized matrix effects, we aimed to separate the analytes of interest after manual sample cleanup using a 2D-LC-setup, combining online solid phase extraction (SPE) with elution on an analytical column.
2. Material and methods
2.1. Chemicals and reagents
Calibrators, quality controls and chromatographic mobile phases for the analytical column were part of the MassTox® TDM A Series from Chromsystems (Gräfelfing, Germany). Drug-free serum was obtained from the blood donation centre of the Bavarian Red Cross (Wiesentheid, Germany).
Reference standards were used for instrument tuning, to establish sample cleanup and chromatography, recovery and quantitative matrix effect testing. Remdesivir (purity 99.6 %) was from Biosynth Carbosynth (Compton, Berkshire, United Kingdom), GS-441524 (purity 98.2 %) from Hycultec (Beutelsbach, Germany). Chloroquine diphosphate (purity 99.3 %), hydroxychloroquine sulfate (purity 99.3 %), ritonavir (purity 98.7 %), lopinavir (purity 97.1 %), and azithromycin dihydrate (purity 92.3 %) were obtained from Supelco (Bellafonte, Pennsylvania, United States). Favipiravir (purity 98.0 %) was from Toronto research chemicals (Toronto, Ontario, Canada). Stable isotope-labelled drugs U-ring-remdesivir-13C6, GS–441524-13C5 and favipiravir-13C1 15N1 were obtained from Alsachim (Straßbourg, Grand Est, France). Chloroquine-D4 phosphate, hydroxychloroquine-D4 sulfate, ritonavir-D6, lopinavir-D8, and azithromycin-13C1D3 were from Toronto research chemicals. UHPLC-grade water, acetonitrile, and methanol were from J.T. Baker (Jackson, TN, USA). UHPLC-grade formic acid was purchased from Biosolve (Dieuze, France). All used chemicals were of the highest purity available from the commercial suppliers.
2.2. Calibrators, quality control samples, and internal standard solution
The commercial COVID19 drug set (Chromsystems) includes 3 calibrator and 2 quality control (QC) levels in lyophilized form. To obtain 6 different calibrators and 4 QCs, lyophilized materials were resuspended in different volumes of water and diluted with drug-free serum if necessary. These suspensions were incubated for 15 min at room temperature (RT) using a vortex shaker (Eppendorf, Hamburg, Germany). All calibrators and QCs were aliquoted to 50 μL and stored at –20 °C for up to 4 weeks, according to the manufacturer’s instructions. Final concentrations of calibrators and QCs are given in Table 1 .
Table 1.
Concentrations of calibrators and quality controls for the analytes in this study.
| Analyte | Calibrators (μg/L) |
Quality controls (μg/L) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | A | B | C | D | |
| Remdesivir | 85.20 | 961.5 | 1282 | 2729 | 4094 | 5458 | 85.20 | 170.4 | 2131 | 4094 |
| GS-441524 | 22.20 | 33.20 | 44.30 | 93.40 | 140.1 | 186.8 | 22.20 | 44.40 | 67.40 | 140.1 |
| Chloroquine | 18.50 | 168.0 | 2240 | 447.0 | 670.5 | 894.0 | 18.50 | 37.00 | 352.0 | 670.5 |
| Hydroxychloroquine | 470.0 | 516.8 | 6890 | 1154 | 1731 | 2308 | 470.0 | 940.0 | 943.0 | 1731 |
| Ritonavir | 95.50 | 2267 | 3023 | 6253 | 9380 | 12510 | 95.50 | 191.0 | 4945 | 9380 |
| Lopinavir | 392.0 | 2336 | 3114 | 6294 | 9441 | 12590 | 392.0 | 784.0 | 4967 | 9441 |
| Favipiravir | 3291 | 3890 | 5186 | 10390 | 15590 | 20790 | 3291 | 6582 | 7987 | 15590 |
| Azithromycin | 42.00 | 195.8 | 261.0 | 432.0 | 648.0 | 864.0 | 42.00 | 84.00 | 357.0 | 648.0 |
Stock solutions of internal standards (ISTD) at a concentration of 0.5 mg/L for chloroquine-D4 phosphate salt, GS–441524-13C5 and 1.0 mg/L for hydroxychloroquine-D4 sulfate, ritonavir-D6, lopinavir-D8, U-ring-remdesivir-13C6, favipiravir-13C1 15N1 and azithromycin-13C1D3 were prepared in methanol, respectively. Dilution of the stock solution in methanol yielded the ISTD working solution with the following concentrations: 251 ng/mL U-ring-remdesivir-13C6, 34.0 ng/mL GS–441524-13C5, 1200 ng/mL favipiravir-13C1 15N1, 63.0 ng/mL chloroquine-D4, 173 ng/mL hydroxychloroquine-D4, 580 ng/mL ritonavir-D6, 294 ng/mL lopinavir-D8, and 43.0 ng/mL azithromycin-13C1D3. ISTD stock and working solution were stably stored at –20 °C.
2.3. Sample preparation
Sample preparation with manual protein precipitation was combined with analysis of the extracts by two-dimensional chromatography, including online solid phase extraction (SPE) (see Chapter 2.4 LC–MS/MS conditions).
First, 200 μL ISTD working solution were added to 50 μL sample (either zero, calibrator, QC or patient serum sample) in 1.5 mL micro tubes (Sarstedt, Nümbrecht, Germany) and thoroughly mixed for 30 s using a vortex shaker (Eppendorf, Hamburg, Germany). A blank calibrator (without analyte, without internal standard) was included in each analytical run and prepared by the addition of 200 μL pure methanol. After centrifugation at 15,000 g for 5 min at RT, 50 μL supernatant were diluted with 950 μL methanol-water (1:1; v/v) in glass vials that were placed into the autosampler (8 °C sample cooling) ready for injection.
2.4. LC–MS/MS conditions
Sample analysis was performed with a 2D Acquity UHPLC system consisting of an autosampler, a column manager, a switching valve, two UHPLC pumps (loading and eluting pump) and a column oven interfaced with a triple quadrupole mass spectrometer Xevo TQ-S (Waters, Milford, MA, USA). Protein precipitation extracts were further cleaned up by online solid phase extraction (SPE), followed by analytical separation on a separate column.
The extract was submitted to an Oasis HLB Direct Connect HP column (30 mm x 2.1 mm, 20 μM, Waters) for online-SPE, while analytical separation was performed with the MassTox® TDM MasterColumn® Series A (Chromsystems). The mobile phases were as follows: • loading pump: phase A1 water and B1 acetonitrile-formic acid (99.9:0.01, v/v); • eluting pump: phase A2 MassTox® TDM Series A mobile phase 1 and phase B2 MassTox® TDM Series A mobile phase 2 (Chromsystems). Between each injection, the autosampler needle and injection port were washed with 500 μL acetonitrile (strong wash) and 600 μL methanol-water (10:90, v/v) (weak wash).
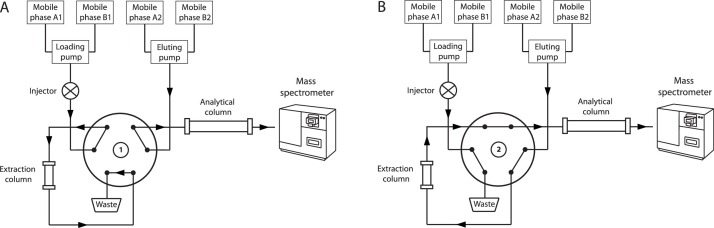
First, 5 μL sample were loaded to the online SPE column with automatic injection mode with phase A1 at a flow rate of 1.5 mL/min. After 1.01 min, the extract was transferred to the analytical column held at 25 °C by the eluting pump delivering mobile phase A2 in back-flush mode, followed by gradient elution with mobile phase A2 and B2. At 2.01 min the switching valve changed back to the starting position and the online SPE column was washed and reconditioned with mobile phase A1 and B1. The elution gradient for the eluting pump with mobile phase B2 was: 0.00–1.50 min → 0 %; 1.51–2.50 min → 50 %; 2.51–3.90 min → 100 %; 3.91–5.00 min → 0 %. The total run time was 5.0 min, whereby the elution entered the mass spectrometer between 1.8 and 3.9 min. The 2D-chromatography configuration and UHPLC program are given in Fig. 1 and Table 2 , respectively.
Fig. 1.
Two-dimensional chromatography configuration. A) Online clean up: The sample protein precipitation extract is pumped to the online SPE column with mobile phase A1 for further extraction. B) Analytic separation: The sample is transferred onto the analytical column by backflush with mobile phase A2 and separated with gradient elution using mobile phase A2 and B2. Meanwhile, the online SPE column is washed and reconditioned with mobile phase A1 and B1.
Table 2.
Solvents delivered for the two-dimensional UHPLC method.
| Loading pump |
Eluting pump |
||||
|---|---|---|---|---|---|
| Time [min] |
Flow [mL/min] |
Mobile phase B1 [%] |
Time [min] |
Flow [mL/min] |
Mobile phase B2 [%] |
| 0.00 | 1.5 | 0 | 0.00 | 0.6 | 0 |
| 1.00 | 1.5 | 0 | 1.50 | 0.6 | 0 |
| 1.01 | 0.1 | 0 | 1.51 | 0.6 | 50 |
| 2.00 | 0.1 | 50 | 2.50 | 0.6 | 50 |
| 2.01 | 1.5 | 50 | 2.51 | 0.6 | 100 |
| 2.50 | 1.5 | 50 | 3.90 | 0.6 | 100 |
| 2.60 | 1.5 | 100 | 3.91 | 0.6 | 0 |
| 3.90 | 1.5 | 100 | 5 | 0.6 | 0 |
| 4.00 | 1.5 | 0 | |||
| 5.00 | 1.5 | 0 | |||
All analytes and corresponding ISTDs were detected with electrospray ionization in the positive mode in multiple reaction monitoring (MRM) to record two mass transitions (m/z), except for favipiravir (M+H, 158.1 m/z) where only a single mass transition was recorded due to a relatively poor fragmentation pattern. The following settings were used: capillary voltage 2.0 kV; source temperature 150 °C; desolvation temperature 450 °C; desolvation gas flow rate, 900 L/Hr; cone gas flow, 150 L/Hr. The MS parameters collision energy, cone voltage and dwell time, were optimized by direct infusion of neat standard solutions with 0.1 μg/mL in methanol. To reduce sensitivity for ritonavir and lopinavir, collision energies were modified for corresponding m/z transitions (detuned). Mass transition-specific detection conditions and analytical retention times are summarized in Table 3 . Analytes were quantified with the TargetLynx V4.1 software (Waters) using the following settings: polynome type, linear; origin, excluded; weighting function, 1/x.
Table 3.
MRM of analytes and corresponding internal standards.
| Compound | Corresponding internal standard | RT [min] |
Precursor ion [M+H] |
Quantifier |
Qualifier |
CV [V] |
Dwell time [s] |
||
|---|---|---|---|---|---|---|---|---|---|
| Product ion [m/z] |
CE [eV] |
Product ion [m/z] |
CE [eV] |
||||||
| Remdesivir | U-ring-remdesivir-13C6 | 3.20 | 603.4 | 402.3 | 14 | 200.1 | 38 | 30 | 0.01 |
| GS-441524 | GS-441524-13C5 | 2.28 | 292.2 | 202.1 | 10 | 163.1 | 25 | 30 | 0.01 |
| Chloroquine | Chloroquine-D4 | 2.26 | 320.2 | 247.2 | 18 | 142.2 | 22 | 30 | 0.01 |
| Hydroxychloroquine | Hydroxchloroquine-D4 | 2.26 | 336.2 | 247.2 | 20 | 158.2 | 20 | 30 | 0.01 |
| Ritonavir | Ritonavir-D6 | 3.37 | 721.3 | 140.2 | 40 | 268.2 | 20 | 30 | 0.01 |
| Lopinavir | Lopinavor-D8 | 3.53 | 629.3 | 155.2 | 25 | 447.3 | 8 | 30 | 0.01 |
| Favipiravir | Favipiravir-13C15N | 2.29 | 158.1 | 85.0 | 20 | – | – | 30 | 0.10 |
| Azithromycin | Azithromycin-13C1D3 | 2.87 | 749.7 | 158.2 | 35 | 591.7 | 25 | 30 | 0.01 |
| U-ring-remdesivir 13C6 | – | 3.20 | 609.3 | 408.4 | 14 | 206.3 | 38 | 30 | 0.01 |
| GS-441524-13C5 | – | 2.28 | 297.3 | 204.1 | 10 | 148.1 | 30 | 30 | 0.01 |
| Chloroquine-D4 | – | 2.26 | 324.3 | 251.2 | 18 | 146.2 | 22 | 30 | 0.01 |
| Hydroxchloroquine-D4 | – | 2.26 | 340.3 | 251.2 | 20 | 162.2 | 20 | 30 | 0.01 |
| Ritonavir-D6 | – | 3.37 | 727.4 | 146.2 | 40 | 274.3 | 20 | 30 | 0.01 |
| Lopinavor-D8 | – | 3.53 | 637.4 | 163.3 | 25 | 447.5 | 8 | 30 | 0.01 |
| Favipiravir-13C115N1 | – | 2.29 | 160.1 | 85.1 | 20 | – | – | 30 | 0.10 |
| Azithromycin-13C1D3 | – | 2.87 | 753.7 | 158.2 | 35 | 595.7 | 25 | 30 | 0.01 |
Retention time, RT; Collision energy, CE; Cone voltage CV.
2.5. Method validation
The ID-LC–MS/MS method was validated on the basis of the guideline of bioanalytical method validation from the European Medicines Agency (EMA), 21 July 2011 [24]. The multi-analyte assay was validated in terms of calibration curve, intra- and inter-assay accuracy and precision, carry-over, selectivity, matrix effect, recovery, dilution integrity, and stability.
2.5.1. Calibration curve
All non-zero calibrators were processed together with a zero calibrator (without analyte, but added internal standard) and a blank (without analyte, without internal standard). Non-zero calibrators should be within ± 15 % of the nominal (theoretical) value, except of the lowest calibrator (termed lower limit of quantification, LLOQ) concentration where it should be within ± 20 %.
2.5.2. Inaccuracy and imprecision
Imprecision was expressed with the coefficient of variation (CV), inaccuracy with relative bias. For all analytes inaccuracy and imprecision were tested by replicate analysis on five different days (5 × 5 design, 5 intra- and 5-inter-day assay) using four different QC samples. The mean concentration should be within ± 15 % of the nominal concentration of the QC samples, except for the LLOQ where it should be within ± 20 %
2.5.3. Carry-over
Carry-over was tested by injection of blank serum samples from different donors after the highest calibrator. The peak area in blanks should not exceed 20 % of the LLOQ peak area and 5 % of the ISTD peak area.
2.5.4. Selectivity
To test for selectivity, leftover anonymized sera from intensive care patients (n = 30), who were not treated with the drugs in this study, but typically with polypharmacy, were investigated. Absence of interfering substances is accepted when the response is ≤ 20 % of the LLOQ for the analytes and ≤ 5 % for the ISTD.
We also assessed the mean ion ratios for quantifiers and qualifiers of the calibrators and ISTD on each day according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI) [25]. For positive samples, the ion ratio of all quantifiers and qualifiers should be ≤ 20 % when compared to the mean ion ratio of corresponding calibrators.
2.5.5. Matrix effects and recovery
Matrix effect testing was done by post-column infusion experiments according to Bonfiglio et al. [26]. Neat analyte and ISTD solutions were directly infused into the mass spectrometer with 0.10 ng/min while processed blank sera (n = 2) were injected to the chromatographic system. Corresponding chromatograms were compared to chromatograms obtained by the injection of pure methanol. Deviations in the baseline or breakdowns of signal intensities would indicate relevant matrix effects.
Quantitative matrix effect and recovery were examined according the EMA guideline [24], and Matuszweski et al. [27] at two different concentrations (low/QC B, high/QC D level). Using the certified reference materials, stock solutions at 1 mg/mL in methanol were prepared for remdesivir, chloroquine, hydroxychloroquine, ritonavir, lopinavir, favipiravir and azithromycin, except for GS-441524 where the solvent was DMSO. Three different sample sets per QC concentration level were compiled, each with identical final analyte and internal standard concentrations in methanol-water (1:1; v/v). Neat analytes and internal standards were directly prepared in methanol-water (1:1; v/v) solvent (set A). Analyte-free hemolyzed, hyperlipidemic and icteric patient sera (n = 6) were spiked with analyte and corresponding internal standards after (set B) and before sample preparation (set C). Matrix effects and recovery were calculated as follows:
| Matrix factor [%] = B/A × 100 |
| Recovery [%] = C/B × 100 |
The internal standard normalized matrix factor (MF) was calculated as the ratio of the MF of the analyte and the MF of the corresponding internal standard and its CV should be ≤ 15 %.
2.5.6. Dilution integrity
To test dilution integrity, a serum sample was prepared with concentrations exceeding the highest calibrator 6 (termed upper limit of quantification, ULOQ) by a factor of x 1.25 for the analytes in this study. Concentrations exceeding the ULOQ were as follows: remdesivir, 6823 μg/L; GS-441524, 233.5 μg/L; chloroquine, 1118 μg/L; hydroxychloroquine, 2885 μg/L; ritonavir, 15640 μg/L; lopinavir, 15740 μg/L; favipiravir, 25990 μg/L; azithromycin, 1080 μg/L. This sample was then diluted 1:5 with drug-free serum in replicate analysis (n = 5). Measured concentrations were back calculated and compared to nominal concentrations, where inaccuracy and imprecision should be within ± 15 %.
2.5.7. Stability
Stability was tested with QC B and QC D samples (refer to Table 1 for concentrations) that were stored up to 4 h at RT (benchtop stability) and -20 °C for four weeks. Autosampler stability of processed samples was also tested for 24 h at 8 °C. Freeze and thaw stability was tested in 3 cycles at -20 °C (freeze time > 12 h) and thawing at room temperature. Stability was tested for each storage condition by measuring QC samples in triplicate using freshly prepared calibration samples. Stability in all conditions was accepted with a deviation of ≤ 15 % from the nominal concentration.
2.6. Application to clinical samples
The validated 2D-UHPLC-MS/MS method was applied to anonymized leftover serum blood samples from patients receiving hydroxychloroquine, ritonavir, azithromycin, or remdesivir (six samples per analyte). Analysis was conducted in accordance with the Declaration of Helsinki and ethical approval was obtained from the local Ethics Committee (document number KB 20/029).
According to the summary of product characteristics and National institute of Health (NIH) COVID19 treatment guidelines [28] dosages are as follows: remdesivir, 200 mg on day 1 and then 100 mg daily for up to 9 days; hydroxychloroquine, 2 × 200−400 mg daily; ritonavir, 2 × 100 mg daily combined with 400 mg lopinavir; azithromycin, 1 × 500 mg daily. Specimens were immediately transferred to the laboratory and sera obtained from collection tubes without gel additive by centrifugation at 2000×g for 10 min at 20 °C. Supernatants were stored up to two weeks at −80 °C until analysis.
3. Results
3.1. Method validation
3.1.1. Calibration curve
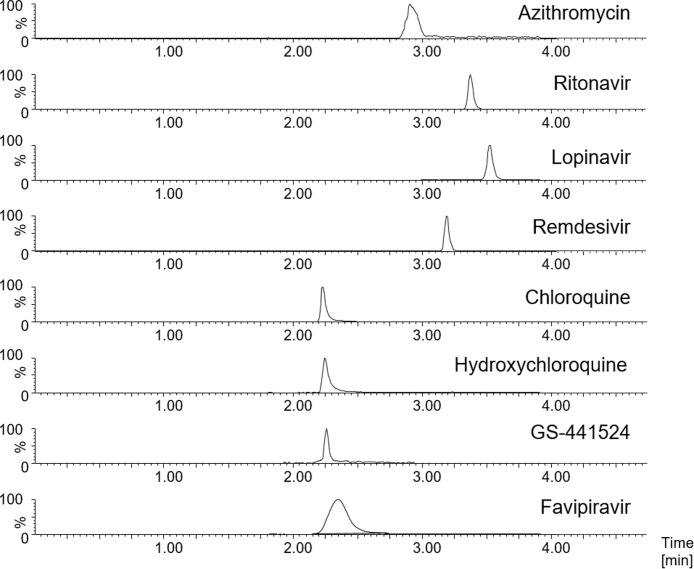
The calibration curve was generated using 6 calibrators with a linear regression model and weighting factor 1/x. The linearity of the method was shown for all calibrations with R2 ≥ 0.993 for all analytes in this study. All calibrators met the EMA specifications with ± 15 % of nominal value, except the LLOQ where it was within ± 20 %. A representative analytical chromatogram is given in Fig. 2 .
Fig. 2.
Representative ID-LC–MS/MS chromatogram of the analytes in this study with MRM acquisition using the lowest calibrator.
3.1.2. Inaccuracy and imprecision
For all analytes intra- and inter-day inaccuracy was ≤ 9.59 %intra- and inter-day imprecision was ≤ 11.1 %, respectively. The respective analytical performance data is summarized in Table 4 .
Table 4.
Intra-day (n = 5) and inter-day (n = 5) inaccuracy and imprecision, matrix effect and recovery results.
| Analyte | QC sample |
Inaccuracy [%] |
Imprecision [%] |
Matrix effect [%] |
Recovery [%] |
||||
|---|---|---|---|---|---|---|---|---|---|
| Intra-day | Inter-day | Intra-day | Inter-day | MFn | CVn | RE | CVr | ||
| Remdesivir | A | 6.82 | 0.78 | 3.29 | 8.93 | / | / | / | / |
| B | 1.20 | 5.36 | 2.58 | 4.49 | 104 | 1.20 | 94.5 | 2.36 | |
| C | 6.93 | 6.24 | 1.66 | 4.19 | / | / | / | / | |
| D | 4.74 | 3.04 | 1.52 | 2.42 | 100 | 1.80 | 100 | 1.69 | |
| GS-441524 | A | 0.98 | 2.57 | 7.38 | 5.35 | / | / | / | / |
| B | 4.13 | 1.50 | 3.54 | 7.83 | 112 | 4.29 | 90.7 | 7.96 | |
| C | 9.39 | 1.48 | 8.18 | 11.1 | / | / | / | / | |
| D | 6.94 | 4.04 | 2.69 | 6.32 | 104 | 6.22 | 101 | 2.50 | |
| Chloroquine | A | 7.05 | 7.80 | 1.74 | 2.39 | / | / | / | / |
| B | 4.40 | 4.49 | 3.63 | 3.50 | 106 | 2.47 | 95.5 | 5.41 | |
| C | 5.27 | 4.06 | 1.76 | 3.30 | / | / | / | / | |
| D | 4.28 | 5.49 | 2.14 | 5.15 | 95 | 0.87 | 96.1 | 7.75 | |
| Hydroxychloroquine | A | 8.89 | 4.02 | 2.23 | 1.60 | / | / | / | / |
| B | 2.74 | 1.93 | 1.38 | 1.55 | 99 | 0.72 | 92.6 | 5.27 | |
| C | 4.24 | 6.22 | 1.88 | 2.00 | / | / | / | / | |
| D | 6.14 | 4.41 | 1.34 | 1.96 | 93 | 0.84 | 95.2 | 5.99 | |
| Ritonavir | A | 0.57 | 4.52 | 1.70 | 1.88 | / | / | / | / |
| B | 5.73 | 6.76 | 2.62 | 1.50 | 99 | 1.12 | 102 | 2.31 | |
| C | 5.74 | 6.61 | 0.74 | 3.43 | / | / | / | / | |
| D | 5.50 | 5.25 | 0.94 | 2.21 | 101 | 0.68 | 107 | 2.05 | |
| Lopinavir | A | 4.72 | 2.18 | 1.00 | 4.44 | / | / | / | / |
| B | 0.86 | 3.76 | 1.26 | 3.09 | 100 | 0.88 | 99.6 | 2.77 | |
| C | 7.64 | 6.85 | 1.44 | 3.23 | / | / | / | / | |
| D | 7.66 | 4.33 | 0.90 | 1.87 | 101 | 0.97 | 104 | 184 | |
| Favipiravir | A | 9.59 | 2.39 | 2.25 | 4.00 | / | / | / | / |
| B | 3.98 | 2.93 | 2.58 | 3.97 | 101 | 2.00 | 97.9 | 1.37 | |
| C | 4.82 | 1.34 | 1.27 | 3.88 | / | / | / | / | |
| D | 9.50 | 4.13 | 3.05 | 5.57 | 103 | 2.68 | 101 | 1.95 | |
| Azithromycin | A | 6.55 | 0.85 | 7.85 | 10.3 | / | / | / | / |
| B | 1.33 | 2.41 | 4.97 | 5.21 | 99 | 3.80 | 86.8 | 3.68 | |
| C | 7.65 | 8.40 | 3.77 | 2.61 | / | / | / | / | |
| D | 5.71 | 8.58 | 4.57 | 3.48 | 102 | 3.90 | 96.3 | 2.98 | |
Internal standard normalized matrix factor, MFn; Coefficient of variation of internal standard normalized matrix factor, CVn,;Recovery, RE; Coefficient of variation of recovery, CVr.
3.1.3. Carry-Over
Peak-areas in blank serum samples injected after the highest calibrator were ≤ 10.8 %, ≤ 8.09 %, ≤ 10.2 %, ≤ 15.8 %, ≤ 2.55 %, ≤ 1.11 %, and ≤ 10.5 % of the lowest calibrator for remdesivir, GS-441524, hydroxychloroquine, ritonavir, lopinavir, favipiravir and azithromycin; except for chloroquine where it was 76.1 %. The peak-area of respective internal standards were consistently ≤ 3.17 %.
3.1.4. Selectivity
The response was ≤ 0.76 %, ≤ 9.59 %, ≤ 8.67 %, ≤ 0.99 %, ≤ 1.49 %, ≤ 0.67 %, ≤ 0.25 % and ≤ 11.5 % of the lowest calibrator for remdesivir, GS-441524, chloroquine, hydroxychloroquine, ritonavir, lopinavir, favipiravir and azithromycin and ≤ 1.17 % for all ISTDs. The mean ion ratios of quantifier to qualifier of analytes and corresponding ISTDs were 1.9 for remdesivir, 1.30 for GS-441524, 2.0 for chloroquine, 3.5 for hydroxychloroquine, 1.0 for ritonavir, 0.72 for lopinavir, and 1.0 for azithromycin. No ion ratio could be established for favipiravir, given that only a single mass transition was recorded.
3.1.5. Matrix effects and recovery
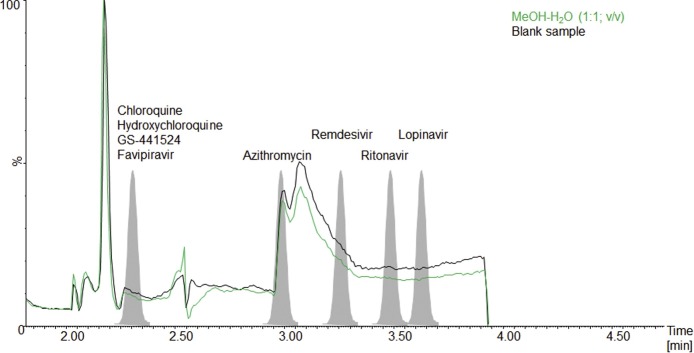
The post-column infusion experiment indicated no relevant matrix effects at the analytic retention time for the analytes and ISTDs in this study given that no noticeable differences were observed between the injection of processed blank sera and pure methanol. The results for qualitative matrix effect testing are shown in Fig. 3 .
Fig. 3.
Post-column infusion experiment for the analytes in this study: Green line, methanol-water (1:1; v/v), black line, processed blank sample.
The CV of the internal standard normalized matrix factor ranged from 0.68 % to 6.22 % and recoveries were between 86.8 % and 107 %. Quantitative matrix effect and recovery results are summarized in Table 4.
3.1.6. Dilution integrity
Dilution integrity was given for all analytes with inaccuracy and imprecision values of ≤ 9.02 % and ≤ 10.8 % for the 1:5 dilution.
3.1.7. Stability
Stability was given for all analytes with deviations ≤ 14.4 % for all conditions tested. Benchtop stability (4 h, RT), autosampler stability (24 h, 8 °C), freeze thaw testing (-20 °C and RT, freeze time > 12 h), and long term stability (4 weeks, -20 °C) gave maximum deviations of 12.1 %, 9.3 %, 12.9 and 14.4 %, respectively.
3.2. Application to clinical samples
Concentrations between 509.4 μg/L and 745.0 μg/L were obtained for hydroxychloroquine, between 398.6 μg/L and 1067 μg/L for ritonavir, between 60.67 μg/L and 174.7 μg/L for GS-441524, and between 64.36 μg/L and 284.6 μg/L for azithromycin, respectively. Remdesivir was < LLOQ in all analyzed samples. The calibration range was appropriate for analysis of the clinical routine specimens.
4. Discussion
We developed and validated a two-dimensional isotope dilution LC–MS/MS method for the simultaneous quantification of remdesivir and its metabolite GS-441524, chloroquine, hydroxychloroquine, ritonavir, lopinavir, favipiravir and azithromycin in human serum.
The crucial advantage of combining two-dimensional chromatography with isotope dilution standardization is that matrix effects can be minimized, which is particularly important in samples from critically ill patients with the risk of unforeseen analytical interference. Furthermore, the use of an online-extraction column contributes to the longevity of the analytical column and avoidance of mass spectrometer contamination by transferring remaining matrix components (in particular peptide/protein residues and salts) directly to the waste. In addition, one-dimensional liquid chromatography is not always capable of efficiently separating analytes of interest in complex samples. However, multidimensional separation methodologies have higher technical requirements when compared to one-dimensional LC-systems.
To quantify all analytes with a uniform injection volume and adequate sensitivity, we performed analyte detuning for ritonavir and lopinavir by adjusting the collision energies. For the quality controls tested, both intra- and inter-day inaccuracy and imprecisions were ≤ 9.59 % and ≤ 11.1, complying with the EMA specifications [24]. Carry-over was negligible for all analytes, except for chloroquine where it was ≈ 76 % of the lowest calibrator. Even though we tested several wash conditions, carry-over could not be reduced any further, probably due to the relatively low chloroquine concentration in the LLOQ and our autosampler instrumentation. To solve this issue, samples with different analytes can be injected in alternating order e.g. chloroquine-remdesivir-chloroquine given that these two drugs are not administered simultaneously in patients. Alternatively, a blank can be injected between consecutive chloroquine measurements. Testing of 30 blank samples from polypharmacized intensive care patients and monitoring of MRM ion ratios for both the analytes and corresponding internal standards confirmed assay selectivity. The method was not affected by hemolysis, icterus, lipemia and the internal standards consistently compensated for matrix effects. Concentrations above the highest calibrator can reliably be quantified by 1:5 dilution using blank serum.
Our stability experiments showed adequate benchtop (4 h, RT), autosampler (24 h, 8 °C) and long-term stability (4 weeks, - 20 °C) for all analytes in this study. According to our freeze-thaw experiments, samples can be repeatedly thawed for analysis. Avataneo et al. reported significant degradation of remdesivir (but not GS-441524) in unextracted plasma, presumably due to the presence of endogenous esterases [10]. In order to prevent the degradation of remdesivir in vitro, corresponding clinical samples should therefore be processed either immediately or after storage of up to four weeks at −20 °C or up to 6 months at −80 °C [10].
We applied the method to anonymized serum samples from patients receiving hydroxychloroquine, ritonavir, azithromycin or remdesivir. Concentrations to be expected from known pharmacokinetic data were obtained within the calibration range of the assay, taking into account the average blood-plasma ratio of 7.2 (± 4.2) for hydroxychloroquine [29]. In agreement with a recently published pharmacokinetic study in two patients receiving treatment with remdesivir [30], the concentration of the prodrug remdesivir (t½, ≈ 1 h) was < LLOQ in our samples, while GS-441524 (t½, ≈ 24 h) was consistently present in corresponding specimens, stressing the necessity for concomitant GS-441524 monitoring.
According to the EMA-guideline on bioanalytical method validation [24], the calibration range of an analytical assay is defined by the calibrators. Samples with concentrations exceeding the highest calibrator, (termed upper limit of quantification, ULOQ) can be diluted into the calibration range, while concentrations below the lowest calibrator (termed lowest limit of quantification, LLOQ) are reported with “< LLOQ”. Depending on the dosage regimens to be subject to TDM and the scientific objective (e.g. pharmacokinetic sampling), the measuring range may be extended with additional calibrators. However, this would typically require a separate partial validation.
In conclusion, we report a robust two-dimensional ID-LC–MS/MS method for the simultaneous quantification of seven repurposed drugs possibly active against COVID-19 in human serum, namely remdesivir (plus metabolite GS-441524), chloroquine, hydroxychloroquine, ritonavir, lopinavir, favipiravir and azithromycin. The assay can be used for routine TDM in clinical laboratories and may help to increase efficacy and safety of repurposed drugs against COVID-19.
Author’s contribution
Katharina Habler and Michael Paal conceptualized the study, developed the method and wrote the manuscript. Uwe Liebchen, Christina Scharf and Ulf Schönermarck helped to implement the method considering clinical aspects. Michael Vogeser, Mathias Brügel und Daniel Teupser supervised the project. All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
Research funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Ethics approval
The Ethical Review Committee of the Ludwig-Maximilians-University Munich approved the study (document number KB 20/029).
CRediT authorship contribution statement
Katharina Habler: Conceptualization, Validation, Writing - original draft, Writing - review & editing. Mathias Brügel: . Daniel Teupser: . Uwe Liebchen: . Christina Scharf: . Ulf Schönermarck: . Michael Vogeser: . Michael Paal: .
Declaration of Competing Interest
The authors have no conflict of interest to declare.
References
- 1.Singh A.K., Majumdar S., Singh R., Misra A. Role of corticosteroid in the management of COVID-19: a systemic review and a Clinician’s perspective. Diabetes Metab. Syndr. 2020;14:971–978. doi: 10.1016/j.dsx.2020.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lythgoe M.P., Middleton P. Ongoing clinical trials for the management of the COVID-19 pandemic. Trends Pharmacol. Sci. 2020;41:363–382. doi: 10.1016/j.tips.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., et al. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., et al. Experimental treatment with Favipiravir for COVID-19: an open-label control study. Engineering (Beijing) 2020 doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meini S., Pagotto A., Longo B., Vendramin I., Pecori D., Tascini C. Role of Lopinavir/Ritonavir in the treatment of Covid-19: a review of current evidence, guideline recommendations, and perspectives. J. Clin. Med. 2020;9 doi: 10.3390/jcm9072050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bleyzac N., Goutelle S., Bourguignon L., Tod M. Azithromycin for COVID-19: more than just an antimicrobial? Clin. Drug Investig. 2020;40:683–686. doi: 10.1007/s40261-020-00933-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pathak D.S.K., Salunke D.A.A., Thivari D.P., Pandey A., Nandy D.K., VKRD Harish, et al. No benefit of hydroxychloroquine in COVID-19: results of systematic review and meta-analysis of randomized controlled trials. Diabetes Metab. Syndr. 2020;14:1673–1680. doi: 10.1016/j.dsx.2020.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuki K., Fujiogi M., Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 2020;215 doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venisse N., Peytavin G., Bouchet S., Gagnieu M.C., Garraffo R., Guilhaumou R., et al. Concerns about pharmacokinetic (PK) and pharmacokinetic-pharmacodynamic (PK-PD) studies in the new therapeutic area of COVID-19 infection. Antiviral Res. 2020;181 doi: 10.1016/j.antiviral.2020.104866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avataneo V., de Nicolo A., Cusato J., Antonucci M., Manca A., Palermiti A., et al. Development and validation of a UHPLC-MS/MS method for quantification of the prodrug remdesivir and its metabolite GS-441524: a tool for clinical pharmacokinetics of SARS-CoV-2/COVID-19 and Ebola virus disease. J. Antimicrob. Chemother. 2020;75:1772–1777. doi: 10.1093/jac/dkaa152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.2020. Summary on Compassionate Use: Remdesivir Gilead.https://www.ema.europa.eu/en/documents/other/summary-compassionate-use-remdesivir-gilead_en.pdf 03 April, Accessed June 2020. [Google Scholar]
- 12.Zuluaga-Idarraga L., Yepes-Jimenez N., Lopez-Cordoba C., Blair-Trujillo S. Validation of a method for the simultaneous quantification of chloroquine, desethylchloroquine and primaquine in plasma by HPLC-DAD. J. Pharm. Biomed. Anal. 2014;95:200–206. doi: 10.1016/j.jpba.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Kaewkhao K., Chotivanich K., Winterberg M., Day N.P., Tarning J., Blessborn D. High sensitivity methods to quantify chloroquine and its metabolite in human blood samples using LC-MS/MS. Bioanalysis. 2019;11:333–347. doi: 10.4155/bio-2018-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morita S., Takahashi T., Yoshida Y., Yokota N. Population pharmacokinetics of hydroxychloroquine in japanese patients with cutaneous or systemic lupus erythematosus. Ther. Drug Monit. 2016;38:259–267. doi: 10.1097/FTD.0000000000000261. [DOI] [PubMed] [Google Scholar]
- 15.Chhonker Y.S., Sleightholm R.L., Li J., Oupicky D., Murry D.J. Simultaneous quantitation of hydroxychloroquine and its metabolites in mouse blood and tissues using LC-ESI-MS/MS: an application for pharmacokinetic studies. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2018;1072:320–327. doi: 10.1016/j.jchromb.2017.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirier J.M., Robidou P., Jaillon P. Simple and simultaneous determination of the hiv-protease inhibitors amprenavir, atazanavir, indinavir, lopinavir, nelfinavir, ritonavir and saquinavir plus M8 nelfinavir metabolite and the nonnucleoside reverse transcriptase inhibitors efavirenz and nevirapine in human plasma by reversed-phase liquid chromatography. Ther. Drug Monit. 2005;27:186–192. doi: 10.1097/01.ftd.0000152680.36517.5d. [DOI] [PubMed] [Google Scholar]
- 17.Dickinson L., Robinson L., Tjia J., Khoo S., Back D. Simultaneous determination of HIV protease inhibitors amprenavir, atazanavir, indinavir, lopinavir, nelfinavir, ritonavir and saquinavir in human plasma by high-performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2005;829:82–90. doi: 10.1016/j.jchromb.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 18.Madelain V., Guedj J., Mentre F., Nguyen T.H., Jacquot F., Oestereich L., et al. Favipiravir pharmacokinetics in nonhuman Primates and insights for future efficacy studies of hemorrhagic fever viruses. Antimicrob. Agents Chemother. 2017:61. doi: 10.1128/AAC.01305-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irie K., Nakagawa A., Fujita H., Tamura R., Eto M., Ikesue H., et al. Pharmacokinetics of Favipiravir in critically ill patients with COVID-19. Clin. Transl. Sci. 2020;13:880–885. doi: 10.1111/cts.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen B.M., Liang Y.Z., Chen X., Liu S.G., Deng F.L., Zhou P. Quantitative determination of azithromycin in human plasma by liquid chromatography-mass spectrometry and its application in a bioequivalence study. J. Pharm. Biomed. Anal. 2006;42:480–487. doi: 10.1016/j.jpba.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 21.Filist M., Bus-Kwasnik K., Ksycinska H., Rudzki P.J. Simplified LC-MS/MS method enabling the determination of azithromycin in human plasma after a low 100mg dose administration. J. Pharm. Biomed. Anal. 2014;100:184–189. doi: 10.1016/j.jpba.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 22.Seger C., Salzmann L. After another decade: LC-MS/MS became routine in clinical diagnostics. Clin. Biochem. 2020;82:2–11. doi: 10.1016/j.clinbiochem.2020.03.004. [DOI] [PubMed] [Google Scholar]
- 23.Iguiniz M., Heinisch S. Two-dimensional liquid chromatography in pharmaceutical analysis. Instrumental aspects, trends and applications. J. Pharm. Biomed. Anal. 2017;145:482–503. doi: 10.1016/j.jpba.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 24.European Medicines Agency . 2011. Guideline on Bioanalytical Method Validation, London, UK.https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-bioanalytical-methodvalidation_en.pdf Accessed January 2020. [DOI] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute . 1st ed. CLSI; Wayne, PA, United States: 2014. Liquid Chromatography-mass Spectrometry Methods; C62-A. [Google Scholar]
- 26.Bonfiglio R., King R.C., Olah T.V., Merkle K. The effects of sample preparation methods on the variability of the electrospray ionization response for model drug compounds. Rapid Commun. Mass Spectrom. 1999;13:1175–1185. doi: 10.1002/(SICI)1097-0231(19990630)13:12<1175::AID-RCM639>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 27.Matuszewski B.K., Constanzer M.L., Chavez-Eng C.M. Strategies for the assessment of matrix effect in quantitative bioanalytical methods based on HPLC-MS/MS. Anal. Chem. 2003;75:3019–3030. doi: 10.1021/ac020361s. [DOI] [PubMed] [Google Scholar]
- 28.COVID-19 Treatment Guidelines Panel. Coronavirus Disease 2019 (COVID-19) Treatment Guidelines. National Institutes of Health. Available at https://www.covid19treatmentguidelines.nih.gov/. Accessed December 2020. [PubMed]
- 29.Tett S.E., Cutler D.J., Day R.O., Brown K.F. A dose-ranging study of the pharmacokinetics of hydroxy-chloroquine following intravenous administration to healthy volunteers. Br. J. Clin. Pharmacol. 1988;26:303–313. doi: 10.1111/j.1365-2125.1988.tb05281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tempestilli M., Caputi P., Avataneo V., Notari S., Forini O., Scorzolini L., et al. Pharmacokinetics of remdesivir and GS-441524 in two critically ill patients who recovered from COVID-19. J. Antimicrob. Chemother. 2020;75:2977–2980. doi: 10.1093/jac/dkaa239. [DOI] [PMC free article] [PubMed] [Google Scholar]