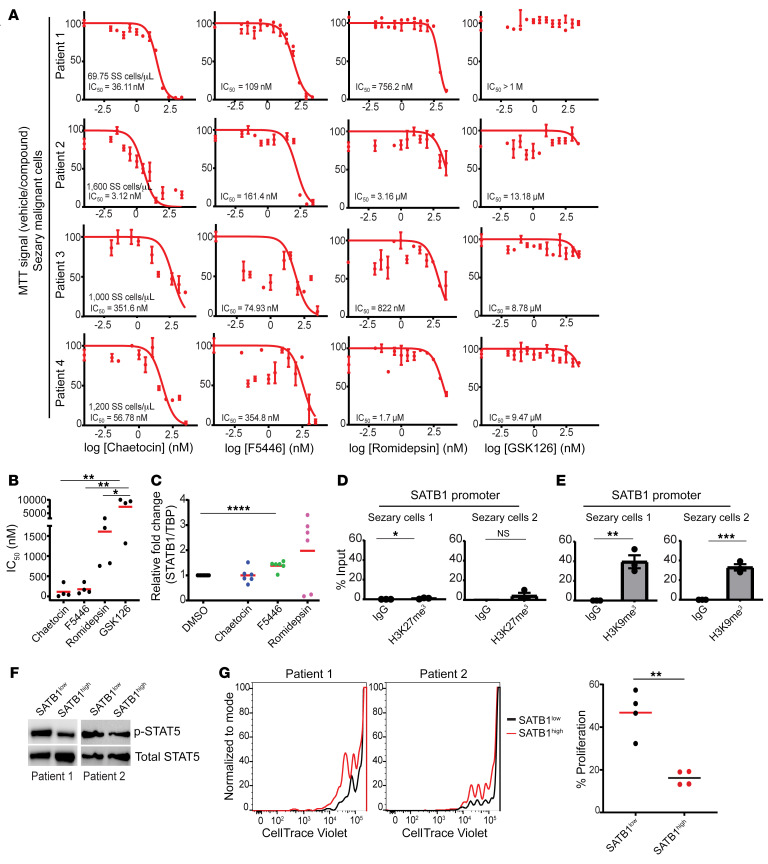
Figure 7. H3K9me3 repression of SATB1 in malignant Sézary patient cells.
(A) CD4+CD26– isolated T cells from peripheral blood apheresis of Sézary patients (n = 4) were cultured in R10 media with 100 U/mL human recombinant IL-2 and treated with increasing doses of SUV39H1/2 inhibitors chaetocin and F5446 (72 hours) as well as romidepsin (72 hours) and GSK126 (48 hours) versus the vehicle control (DMSO) prior to MTT analysis. IC50 values (nM) were calculated for each patient sample for the respective treatments. (B) Summary of IC50 values for chaetocin, F5446, romidepsin, and GSK126 for each malignant sample (n = 4). One-way ANOVA with Tukey’s multiple-comparison test: *P < 0.05; **P ≤ 0.01. (C) RNA was extracted from primary CD4+CD26– Sézary patient cells that were treated with chaetocin, F5446, romidepsin, or vehicle control (DMSO) for 24–36 hours, and SATB1 mRNA expression was quantified by q-PCR normalized to human TBP mRNA. Data pooled from 3 patient samples with 2 independent experiments shown (n = 6). Two-tailed Student’s t test: ****P ≤ 0.0001. (D) Chromatin immunoprecipitation quantified by real-time q-PCR with anti-H3K27me3 (clone C36B11) or control IgG isotype immunoprecipitation from isolated CD4+ T cells from peripheral blood of Sézary patients (n = 2) calculated against 2.5% input values. Regions amplified at the predicted occupied region (approximately 4.8 kb)of the SATB1 promoter. Representative of 2 independent experiments. Two-tailed Student’s t test: *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001. (E) Similar to D except with anti-H3K9me3 (Abcam, ab8898) at the approximately 5.6-kb region versus the control region (n = 2). Representative of 2 independent experiments. Two-tailed Student’s t test: *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001. (F) Primary Sézary CD4+CD26– cells (n = 2) were transduced with retrovirus containing human SATB1 and sorted for GFP+ and GFP– cells. Western blot was performed with antibodies against human p-STAT5 and STAT5 protein in cells endogenously expressing SATB1 versus cells with ectopic expression of SATB1 (n = 2). (G) Primary Sézary cells were labeled with CellTrace Violet and were serum starved for 24 hours prior to stimulation with anti-CD3/anti-CD28 beads and cultured in complete medium with 100 U/mL rhIL-2 for 5 days. Proliferation was assessed by CellTrace Violet dilution using FACS for cells ectopically (n = 2) or endogenously (n = 2) expressing SATB1. Experiment was performed on 2 replicates per patient (n = 4). Two-tailed Student’s t test: **P ≤ 0.01.