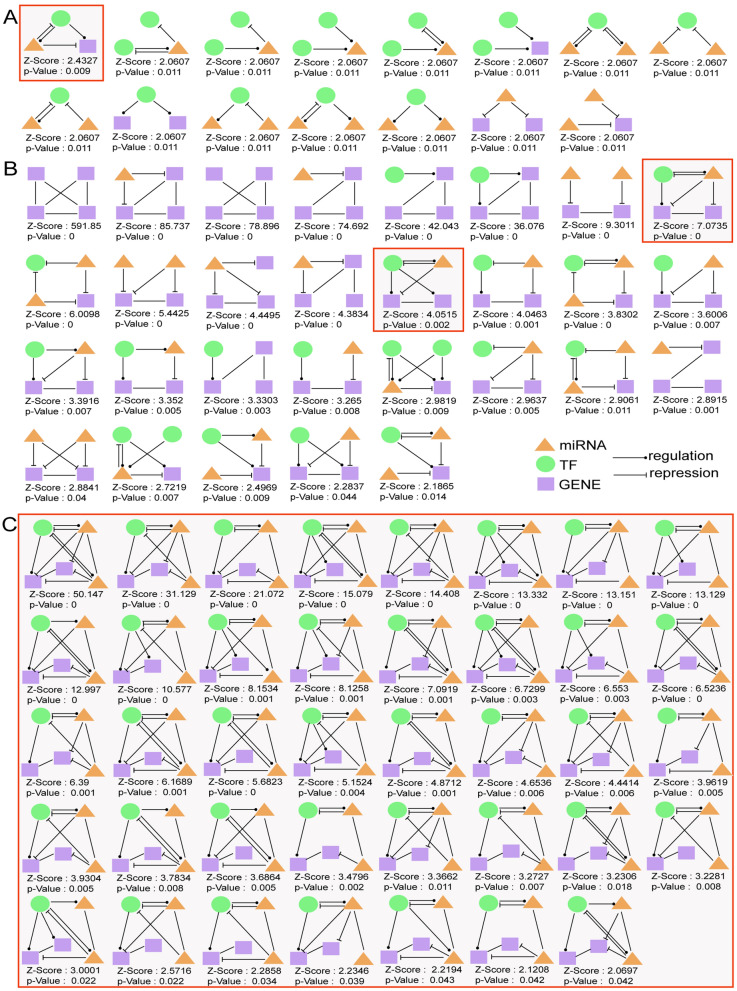
Figure 2.
3-node, 4-node and 5 node motifs. (A) 15 different types of 3-node regulatory motifs. (B) 29 different types of 4-node regulatory motifs. (C) 39 screened different types of 5-node regulatory motifs. The motifs are composed of miRNAs, TFs, and target genes and their Z-scores and p value s are presented. Orange triangles represent miRNAs, green rounds represent TFs and purple squares represent genes. An arrow ending in a circle represents the regulatory relationship, while, an arrow ending in a “T” represents the repression relationship. Six types of relationships involved in these motifs: miRNA–gene (miRNA represses gene expression); miRNA–TF (miRNA represses TF gene expression); TF–miRNA (TF regulates miRNA expression); TF–gene (TF regulates gene expression); gene–gene (gene and gene interaction); and miRNA–miRNA (miRNA and miRNA interaction). Motifs surrounded by red squares (3-node, 4-node and 5-node composite feed-forward loop) as significant motifs were merged to form 3-node, 4-node and 5-node regulatory sub-networks, respectively. TF: transcription factor; FFL: feed-forward loop.