Abstract
More and more studies have reported that dysregulation of microRNAs (miRNAs) leads to the proliferation and EMT of multiple cancers. Recently, several reports have demonstrated that dysregulation of miR-4262 occurs in numerous cancers. However, its role and precise mechanism in human cervical cancer (CC) have not been well clarified. Hence, this study aimed to explore the biological roles and precise mechanisms of miR-4262 in CC cell lines. The level of miR-4262 was found to be significantly decreased in CC tissues and cell lines. Moreover, decreased expression of miR-4262 was closely related to increased expression of Kaiso (ZBTB33), which belongs to the BTB/POZ family, in CC tissues and cell lines. The proliferation and EMT of CC cells were inhibited by a miR-4262 mimic. However, downregulation of miR-4262 enhanced the proliferation and EMT of CC cells. Next, bioinformatics analysis predicted that miR-4262 might directly target the Kaiso gene. Besides, luciferase reporter assay had confirmed this result. Moreover, introduction of Kaiso in CC cells partially blocked the effects of miR-4262 mimic. In conclusion, miR-4262 suppressed the proliferation and EMT of CC cells by directly downregulating Kaiso.
Key words: Cervical cancer (CC), Proliferation, Epithelial–mesenchymal transition (EMT), MicroRNA-4262, Kaiso
INTRODUCTION
Cervical cancer (CC) is the third leading cause of cancer-related deaths among women worldwide1. About 500,000 new cases are diagnosed, with an estimated 300,000 deaths per year1. With advances in surgery, radiotherapy, and chemotherapy, the mortality of CC has decreased2. Because of tumor recurrence and metastasis, patients with advanced CC still have very poor prognosis and significantly variable clinical outcomes3; however, the precise molecular mechanisms of CC are still unclear. Therefore, uncovering the molecular mechanisms of CC and identifying effective biomarkers in the development and progression of CC may provide the available targets for the prevention and treatment of CC.
MicroRNAs (miRNAs) are highly conserved small (about 22 nucleotides in length) noncoding RNAs4,5, which are involved in multiple biologic processes such as proliferation, differentiation, invasion, and apoptosis. miRNAs can downregulate protein expression either by mRNA degradation and/or by translational repression, by recognizing complementary sequences in the 3′-untranslated regions (3′-UTRs) of targeted mRNAs6,7. Increasing evidence has shown that miRNAs play critical roles in various kinds of cancers4–10. It has been reported that miRNAs can be tumor suppressors or oncogenes in CC, depending on the function of their target genes, including miR-5439, miR-94410, miR-3156-3p11, miR-92a12, miR-211113, and miR-42914. miR-4246 has attracted attention because the level of miR-4262 is significantly downregulated, and it functions as a tumor promoter in breast cancer, cutaneous malignant melanoma, and hepatocellular carcinoma15–17, whereas it functions as a tumor suppressor, and its level is significantly upregulated in osteosarcoma18. However, the function and precise molecular mechanism of miR-4262 in CC remain unclear. Moreover, Kaiso, also called ZBTB33, is a transcriptional repressor and belongs to the BTB/pox virus and zinc finger (POZ) family19,20. More and more evidence demonstrates that Kaiso is involved in many biologic processes such as cell proliferation, invasion, and epithelial–mesenchymal transition (EMT)21–23. Kaiso functions as a tumor promoter, is significantly increased in several cancers, and is closely associated with the development and progression of cancers24,25. However, the mechanisms underlying the regulation of Kaiso expression in CC are still unclear.
In this study, frequent downregulation of miR-4262 was detected in CC tissues and cell lines. Upregulation of miR-4262 suppressed the proliferation and EMT of CC cells. Furthermore, the oncogene Kaiso was the direct target of miR-4262. Overexpression of Kaiso blocked the inhibitory effects of miR-4262 on the proliferation and EMT of CC cells. Therefore, these results confirmed important roles for miR-4262 in the pathogenesis of CC, which indicated its potential application in CC treatment.
MATERIALS AND METHODS
Cell Culture and Human Tissue
End1/E6E7, a human normal cervical epithelium cell line, was cultured in keratinocyte serum-free medium (Gibco, Grand Island, NY, USA) containing 0.05 mg/ml bovine pituitary extract, 0.1 ng/ml human recombinant epithelial growth factor (R&D, Minneapolis, MN, USA), and 1% streptomycin–penicillin (Gibco). CC cells such as HT-3, C33A, HeLa229, HeLa, MS751, HCC94, SiHa, CaSKi, and ME-180 were grown in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) containing 10% fetal bovine serum (FBS; Gibco) and 1% streptomycin–penicillin at 37°C in 5% CO2. Ten pairs of fresh CC tissues and their matched adjacent normal tissues were collected after curative surgical operations from Cangzhou Central Hospital (Hebei, P.R. China). All tissue biopsies were immediately frozen in liquid nitrogen until further use. Informed consent was obtained from all patients before surgery, and the study protocol was approved by the Ethics Committee of Cangzhou Central Hospital.
Bioinformatics Analysis of Potential Binding Partners for miR-4262
The bioinformatics program, TargetScan 6.2 (www.targetscan.org), was used to identify putative binding partners for miR-4262.
miRNA Transfection
The miR-4262 mimic, miR-negative control (miR-NC), miR-4262 inhibitor, miR-negative control of inhibitor (NC-inhibitor), the pcDNA3.1 vector, and pcDNA-Kaiso were synthesized from GeneChem (Shanghai, P.R. China). HeLa and CaSKi cells were transfected using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocols.
RNA Extraction and Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from HeLa and CaSKi cells by TRIzol reagent (Invitrogen). For miR-4262 detection, cDNA was synthesized using the PrimeScript RT Reagent Kit (TaKaRa, Japan). The qRT-PCR was performed using SYBR Premix ExTaq (TaKaRa) with the Bio-Rad Real-Time PCR System (Bio-Rad, Hercules, CA, USA). The endogenous small nuclear RNA (snRNA) U6 was used as a housekeeping gene to normalize the level of miR-4262. The relative level of miR-4262 was quantified with the 2−ΔΔCt method. For proliferating cell nuclear antigen (PCNA), cyclin-dependent kinase 4 (CDK4), cyclin D1, cyclin-dependent kinase inhibitor 1A (p21), epithelial cadherin (E-cadherin), vimentin, β-catenin, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA analyses, Moloney murine leukemia virus (M-MLV) Reverse Transcriptase (Promega, Madison, WI, USA) was used for cDNA synthesis. The mRNA level of other genes was normalized to the mRNA expression of GAPDH and was also calculated by the 2−ΔΔCt method. Each sample was assessed in triplicate. Primers used are listed in Table 1.
Table 1.
Sequence of Primers for qRT-PCR
| Gene | Primer Sequence |
|---|---|
| PCNA | F: 5′-CCTGCTGGGATATTAGCTCCA-3′ |
| R: 5′-CAGCGGTAGGTGTCGAAGC-3′ | |
| CDK4 | F: 5′-GGGGACCTAGAGCAACTTACT-3′ |
| R: 5′-CAGCGCAGTCCTTCCAAAT-3′ | |
| Cyclin D1 | F: 5′-GCTGCGAAGTGGAAACCATC-3′ |
| R: 5′-CCTCCTTCTGCACACATTTGAA-3′ | |
| p21 | F: 5′-GTGAAAACAGAGCGAGAGAGATG-3′ |
| R: 5′-CAGGGGTACAGTGCTAAAGGC-3′ | |
| E-cadherin | F: 5′-TACACTGCCCAGGAGCCAGA-3′ |
| R: 5′-TGGCACCAGTGTCCGGATTA-3′ | |
| β-catenin | F: 5′-AAAGCGGCTGTTAGTCACTGG-3′ |
| R: 5′-CGAGTCATTGCATACTGTCCAT-3′ | |
| Vimentin | F: 5′-GACGCCATCAACACCGAGTT-3′ |
| R: 5′-CTTTGTCGTTGGTTAGCTGGT-3′ | |
| U6 | F: 5′-CTCGCTTCGGCAGCACA-3′ |
| F: 5′-AACGCTTCACGAATTTGCGT-3′ | |
| GAPDH | F: 5′-GAGTCAACGGATTTGGTCGTATTG-3′ |
| R: 5′-CCTGGAAGATGGTGATGGGATT-3′ |
PCNA, proliferating cell nuclear antigen; CDK4, cyclin-dependent kinase 4; p21, cyclin-dependent kinase inhibitor 1A; E-cadherin, epithelial cadherin; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Cell Counting Kit-8 Assay
The CCK-8 assay (Dojindo, Japan) was used to detect the viabilities of HeLa and CaSKi cells. HeLa and CaSKi cells were seeded at a density of 1 × 104 cells/well in 96-well plates overnight. Cells were then transfected with miR-4262 mimic and inhibitor for 48 h. The cells were cultured in complete medium containing WST-8 substrate at 37°C for 2 h. The absorbance at 450 nm was detected using an electroluminescence immunosorbent assay reader.
Enzyme-Linked Immunosorbent Assay-Bromodeoxyuridine Assay (ELISA-BrdU Assay)
To investigate the effect of miR-4262 on the proliferation of HeLa and CaSKi cells, cells were seeded at a density of 1 × 104 cells/well in 96-well plates overnight. The cells were transfected with miR-4262 mimic or inhibitor for 48 h at 37°C. Cell proliferation was estimated using Cell Proliferation ELISA-BrdU Kit (Roche Diagnostics, Indianapolis, IN, USA) according to the manufacturer’s instructions.
Western Blot Analysis
The protein was extracted using RIPA lysis buffer (Beyotime Biotechnology, P.R. China) containing protease inhibitors (Millipore, Billerica, MA, USA). The proteins were quantified using the BCA Protein Assay Kit (Beyotime Biotechnology). The Western blot system was established using a Bio-Rad Bis-Tris Gel System according to the manufacturer’s instructions. Primary antibodies against Kaiso (1:1,000; Abcam, Cambridge, MA, USA), p21, cyclin D1, CDK4, PCNA (1:1,000; Cell Signaling Technology, Danvers, MA, USA), and α-tubulin (1:2,000; Sigma-Aldrich, St. Louis, MO, USA) were incubated with the polyvinylidene difluoride (PVDF) membrane (Millipore) at 4°C overnight, followed by washing three times in Tris-buffered saline-Tween (TBST) and incubation with secondary antibodies tagged by horseradish peroxidase (1:1,000; Cell Signaling Technology) for 2 h at room temperature. After rinsing, the protein levels were detected with an enhanced chemiluminescence (ECL) kit (Millipore) following the manufacturer’s instructions.
Dual-Luciferase Reporter Assay
The predicted 3′-UTR sequence of Kaiso interacting with miR-4262 and mutated sequences within the predicted target sites were synthesized and then introduced into the pGL3 vector (Promega). HeLa and CaSKi cells were transfected with miR-4262 mimic or miR-NC, followed by cotransfection with the wild-type or mutant 3′-UTR of Kaiso using FuGENE (Promega). Dual-luciferase reporter assay was carried out on extracts from the cells 48 h after transfection and measured using the Dual-Luciferase Assay System (Promega). The firefly luciferase activity was normalized to the Renilla luciferase activity of each transfected well.
Statistical Analysis
All experiments were repeated three times. The data of multiple experiments are expressed as the mean ± standard error of the mean (SEM). Statistical analyses were performed using GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). One-way ANOVA and Student’s t-test were used to measure the differences between the groups. A value of p < 0.05 was considered as a statistically significant result.
RESULTS
The Level of miR-4262 Was Decreased and the Kaiso Expression Was Increased in CC Tissues and Cell Lines
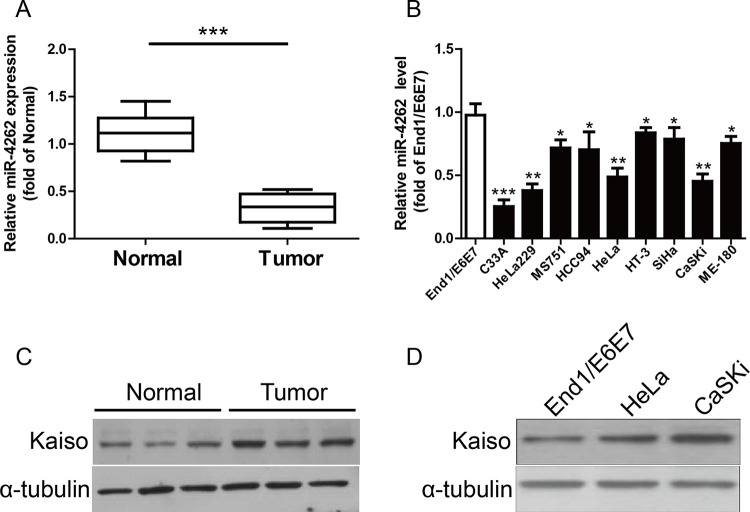
In this study, the miR-4262 level in the CC tissues was significantly decreased compared with normal tissues (Fig. 1A). Next, the level of miR-4262 was estimated by qRT-PCR in several CC cell lines including HT-3, C33A, HeLa229, HeLa, MS751, HCC94, SiHa, CaSKi, ME-180, and a human normal cervical epithelium cell line (End1/E6E7). miR-4262 was significantly downregulated in these CC cell lines compared to that in End1/E6E7 cells (Fig. 1B). Moreover, TargetScan 6.2, an online database, suggested that Kaiso was predicted to be a direct target of miR-4262. For further study, the protein expression of Kaiso in both CC tissues and cell lines was determined and shown to be markedly increased in CC tissues and cell lines (Fig. 1C and D).
Figure 1.
The levels of microRNA-4262 (miR-4262) and Kaiso in cervical cancer (CC) tissues and cell lines. (A) Relative miR-4262 level in CC tissues and their corresponding adjacent normal tissues. (B) Relative miR-4262 level analyzed by qualitative real-time reverse transcription polymerase chain reaction (qRT-PCR) in several CC cell lines (C33A, HeLa229, MS751, HCC94, HeLa, HT-3, SiHa, CaSKi, and ME-180) and a human normal cervical epithelium cell line (End1/E6E7) were normalized with U6 small nuclear RNA (snRNA). (C) The protein level of Kaiso was determined by Western blot in CC tissues. (D) The expression of Kaiso was detected by Western blot in CC cell lines. All data are presented as mean ± standard error of the mean (SEM), n = 6. *p < 0.05, **p < 0.01, ***p < 0.001 versus normal tissues or End1/E6E7.
Effects of miR-4262 on the Proliferation of HeLa and CaSKi Cells
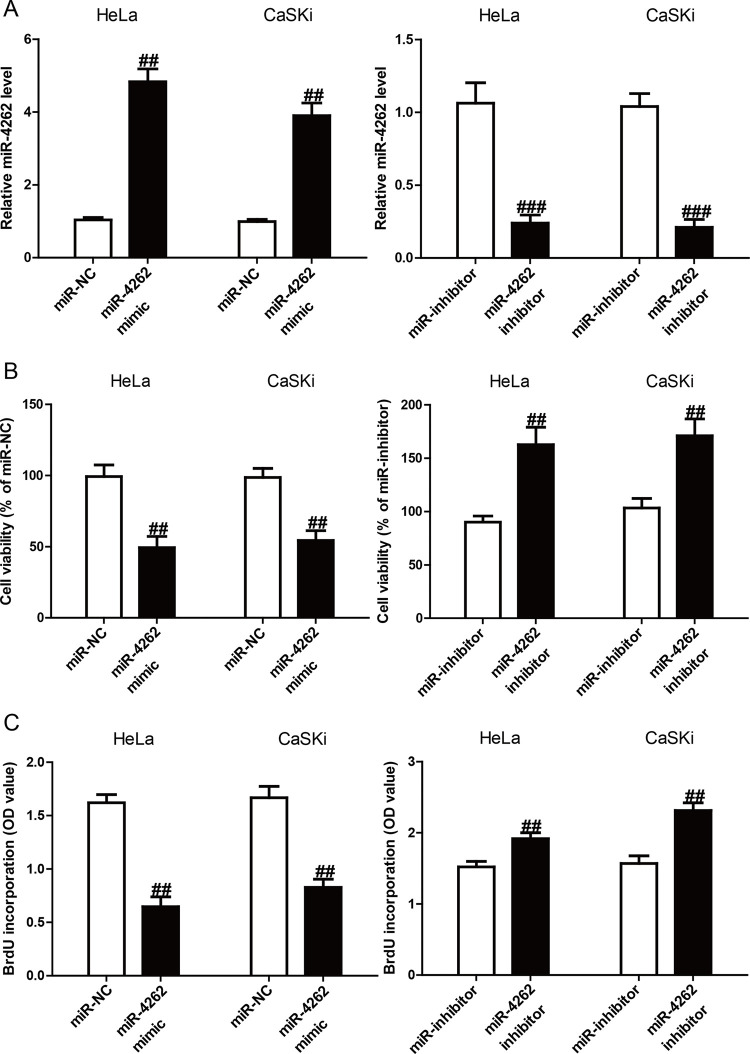
Since miR-4262 was downregulated in CC cells, it was predicted that miR-4262 might function as a tumor suppressor in CC. The miR-4262 level was higher or lower in the miR-4262 mimic or inhibitor group than in the miR-NC or miR-inhibitor group, respectively (Fig. 2A). For detecting the function of miR-4262 in the viability of CC cells, HeLa and CaSKi cells were transfected with miR-4262 mimic and inhibitor. The CCK-8 assay results demonstrated that introduction of miR-4262 significantly reduced the viabilities of HeLa and CaSKi cells, and knockdown of miR-4262 dramatically enhanced the viabilities of both cell lines (Fig. 2B). Furthermore, using the ELISA-BrdU assay, it was demonstrated that introduction of miR-4262 could inhibit the proliferation of both HeLa and CaSKi cells, whereas downregulation of miR-4262 promoted the proliferation of CC cells (Fig. 2C).
Figure 2.
Effects of miR-4262 on cell viabilities and proliferation in CC cells. HeLa and CaSKi cells were transfected with miR-4262 mimic or miR-NC for 48 h. (A) The level of miR-4262 in HeLa and CaSKi cells was determined by qRT-PCR. (B) Cell viability was assessed by the cell counting kit-8 (CCK-8). (C) Cell proliferation was assessed by enzyme-linked immunosorbent assay-bromodeoxyuridine (ELISA-BrdU) assay. All data are presented as mean ± SEM, n = 6. ##p < 0.01, ###p < 0.001 versus miR-NC or miR-inhibitor.
The Effects of miR-4262 on the Expression of Cell Proliferation and Cell Cycle-Related Proteins in CC Cells
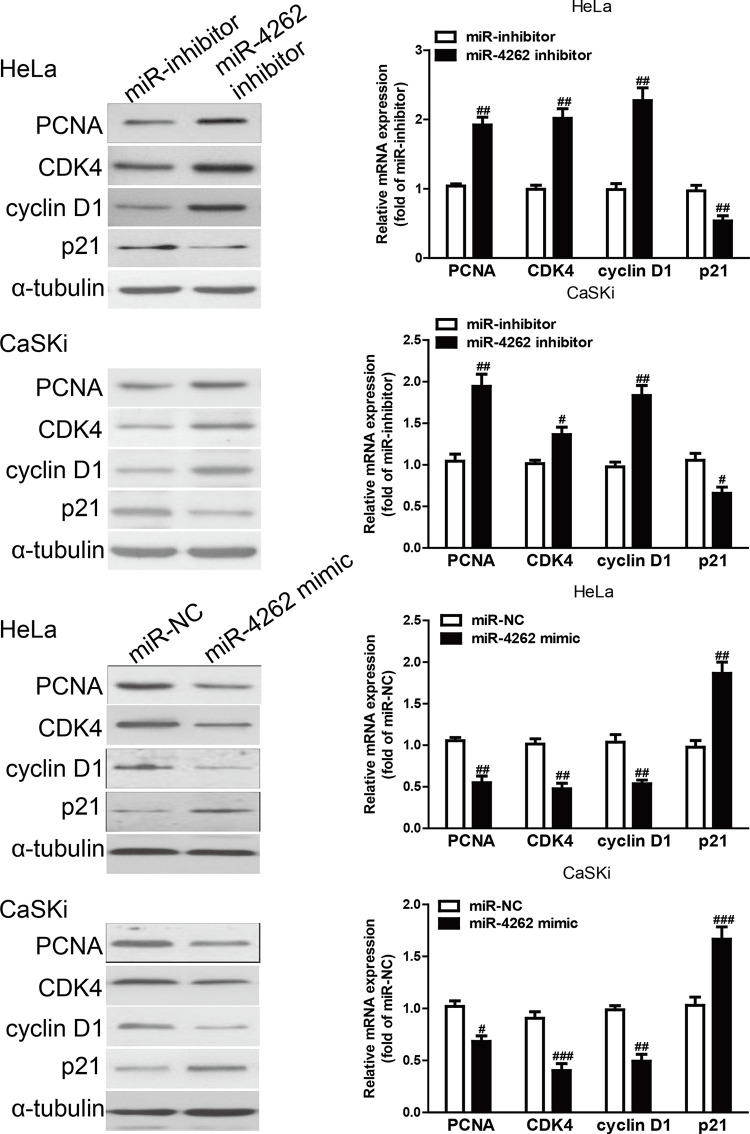
Since miR-4262 could regulate the proliferation of HeLa and CaSKi cells, protein and mRNA expressions of cell proliferation and cell cycle-related genes such as PCNA, CDK4, cyclin D1, and p21 were determined by Western blot analysis and qRT-PCR, respectively. The results showed that the protein and mRNA expressions of PCNA, CDK4, and cyclin D1 were obviously downregulated in the miR-4262 mimic group compared with the miR-NC group, and were significantly upregulated in the miR-4262 inhibitor group compared with the miR-inhibitor group (Fig. 3). Conversely, p21 protein expression was markedly increased by miR-4262 overexpression and decreased by miR-4262 knockdown (Fig. 3). The above data indicated that introduction of miR-4262 might be closely associated with downregulation of PCNA, CDK4, and cyclin D1, and upregulation of p21 in HeLa and CaSKi cells.
Figure 3.
The effects of miR-4262 on the expression of cell proliferation and cell cycle-related proteins in CC cells. HeLa and CaSKi cells were transfected with miR-4262 mimic or inhibitor for 48 h. The protein and mRNA expressions of proliferating cell nuclear antigen (PCNA), cyclin-dependent kinase inhibitor 1A (p21), cyclin D1, and cyclin-dependent kinase 4 (CDK4) in HeLa and CaSKi cells were determined by Western blot and qRT-PCR, respectively. α-Tubulin was detected as a loading control. All data are presented as mean ± SEM, n = 6. #p < 0.05, ##p < 0.01, ###p < 0.001 versus miR-NC or miR-inhibitor.
Effects of miR-4262 on EMT of CC Cells
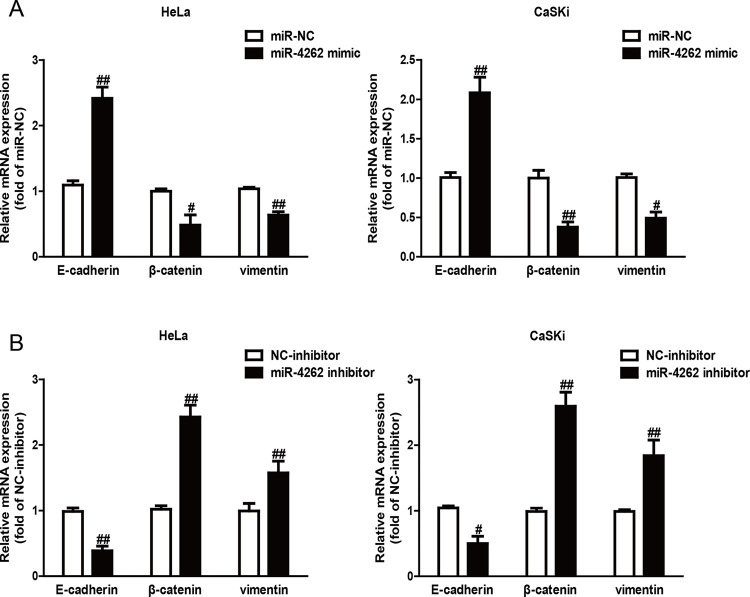
Next, the effects of miR-4262 on the expression of EMT markers in both HeLa and CaSKi cells were explored using qRT-PCR assay. Introduction of miR-4262 in HeLa and CaSKi cells led to increasing expression of the epithelial marker E-cadherin and decreasing expressions of mesenchymal markers vimentin and β-catenin at the mRNA level (Fig. 4A). However, knockdown of miR-4262 had the opposite effects on EMT of CC cells (Fig. 4B). Altogether, the findings revealed that introduction of miR-4262 suppressed EMT of CC cells.
Figure 4.
Overexpression of miR-4262 suppressed epithelial–mesenchymal transition (EMT) of CC cells. HeLa and CaSKi cells were transfected with miR-4262 mimic (A) or inhibitor (B) for 48 h. The expressions of epithelial cadherin (E-cadherin), β-catenin, and vimentin were determined by qRT-PCR in both cell lines. All data are presented as mean ± SEM, n = 6. #p < 0.05, ##p < 0.01 versus miR-NC or miR-inhibitor.
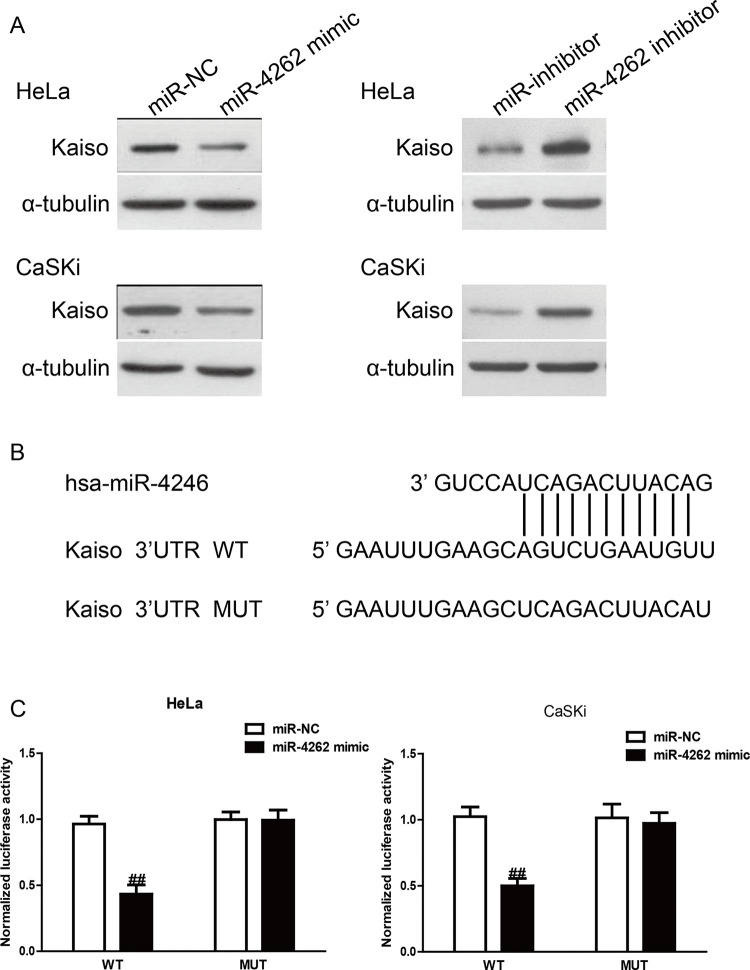
Kaiso Is a Direct Target of miR-4262 in CC Cells
Based on the bioinformatics tool (TargetScan 6.2), the Kaiso gene was identified as a binding target of miR-4262. First, Western blotting was performed to determine the protein expression of Kaiso in HeLa and CaSKi cells after transfection with miR-4262 mimic or inhibitor. As shown in Figure 5A, the protein level of Kaiso was decreased by miR-4246 overexpression in HeLa and CaSKi cells. Moreover, knockdown of miR-4246 increased the protein level of Kaiso. To confirm whether the effects of miR-4262 were mediated by direct binding of miR-4262 with the 3′-UTR of the Kaiso gene, the wild-type 3′-UTR of Kaiso was cloned into the firefly luciferase reporter vector pGL3 (pGL3-Kaiso 3′-UTR WT), and then cotransfected with miR-4246 mimic or miR-NC into HeLa and CaSKi cells for 48 h. Dual-luciferase reporter assays showed that miR-4262 significantly suppressed WT Kaiso-3′-UTR luciferase activity (Fig. 5C). A luciferase reporter plasmid carrying the mutated 3′-UTR of Kaiso (MUT Kaiso-3′-UTR) based on the predicted miR-4246 binding sites (Fig. 5B) was generated and cotransfected. As shown in Figure 5C, miR-4246 had no effect on the activity of luciferase carrying MUT Kaiso-3′-UTR, which suggested that miR-4246 may target the 3′-UTR of Kaiso, resulting in either the degradation or the inhibition of translation of Kaiso relative to miR-NC. These data indicated that the binding of miR-4246 with the 3′-UTR of Kaiso gene was required to produce its effects.
Figure 5.
Kaiso was a direct target of miR-4262. HeLa and CaSKi cells were transfected with miR-4262 mimic or inhibitor for 48 h. (A) The protein expression of Kaiso was determined by Western blot. α-Tubulin was detected as a loading control. (B) Schematic representation of Kaiso 3′-untranslated regions (3′-UTRs) showing putative miRNA target site. (C) The analysis of the relative luciferase activities of wild type (Kaiso-WT) and mutant (Kaiso-MUT) in CC cells. All data are presented as mean ± SEM, n = 6. ##p < 0.01 versus miR-NC.
Overexpression of Kaiso Partially Blocked the Effects of miR-4262 Mimic on the Proliferation and EMT of CC Cells
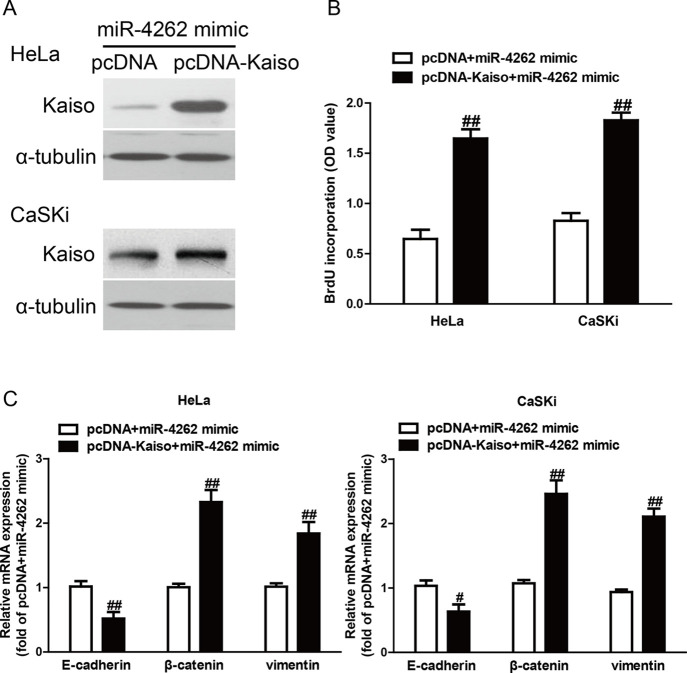
To confirm whether miR-4262 overexpression suppressed the proliferation and EMT of CC cells through downregulation of Kaiso, HeLa and CaSKi cells were cotransfected with miR-4262 mimic and pcDNA-Kaiso (Fig. 6A). The ELISA-BrdU assay revealed that overexpression of Kaiso significantly promoted the proliferation of CC cells transfected with the miR-4262 mimic (Fig. 6B). Moreover, introduction of Kaiso decreased the E-cadherin expression and increased the vimentin and β-catenin expression in HeLa and CaSKi cells after transfection with miR-4262 mimic (Fig. 6C). Hence, the effects of miR-4262 mimic on the proliferation and EMT of CC cells were partially reversed by overexpression of Kaiso. All of these results clearly demonstrated that miR-4262 overexpression suppressed the proliferation and EMT in CC cells possibly by downregulation of Kaiso expression, suggesting that knockdown of Kaiso may be required for the effects of a miR-4262 mimic on the proliferation and EMT of CC cells.
Figure 6.
Overexpression of Kaiso partially rescued miR-4262-inhibited cell proliferation and EMT in CC cells. HeLa and CaSKi cells were transfected with miR-4262 mimic with or without pcDNA-Kaiso. (A) The expression of Kaiso was determined by Western blot. α-Tubulin was also detected as a loading control. (B) Cell proliferation was assessed by ELISA-BrdU assay. (C) The expressions of E-cadherin, β-catenin, and vimentin were determined by qRT-PCR. All data are presented as mean ± SEM, n = 6. #p < 0.05, ##p < 0.01 versus pcDNA + miR-4262 mimic.
DISCUSSION
Previous studies have reported that many miRNAs are dysregulated in multiple types of cancers, and they can contribute to the development and progression of cancers26 as either tumor suppressors or promoters27. miR-4262 has been investigated in hepatocellular carcinoma, cutaneous malignant melanoma, osteosarcoma, and breast cancer15–18. All these previous findings suggest that miR-4262 functions as either a tumor suppressor or a promoter. However, the functional role and precise molecular mechanism of miR-4262 in CC remained unknown. Therefore, this study was designed to confirm the biological function and precise mechanism of miR-4262 in CC. For the first time, it was shown that the miR-4262 level was frequently downregulated in CC tissues and cell lines. According to these outcomes, it is speculated that miR-4262 might be a tumor suppressor in CC. As expected, knockdown of miR-4262 inhibited the proliferation and EMT of HeLa and CaSKi cells. Results suggested that miR-4262 played critical roles in the regulation of the proliferation and EMT of CC cells, and it might therefore be a potential diagnostic and predictive biomarker.
Next, the precise mechanism of miR-4262 was investigated with regard to inhibiting the proliferation and EMT of CC cells. The qRT-PCR, Western blotting, and luciferase reporter assay showed that miR-4262 could directly target the Kaiso gene. Importantly, it also demonstrated that the effects of miR-4262 mimic on the proliferation and EMT of CC cells were partly reversed by the upregulation of Kaiso. This confirmed that miR-4262 played important roles in suppressing the proliferation and EMT of CC cells, partly via downregulation of Kaiso.
In the current study, the results of the CCK-8 and ELISA-BrdU assays showed that overexpression of miR-4262 markedly suppressed the viabilities and proliferation of HeLa and CaSKi cells. Recent reports have demonstrated that a common characteristic of human cancer is disorder of cell cycle regulators, suggesting that a possible way for treating human cancers is the modulation of cell cycle progression in cancer cells28–30. To confirm the potential mechanisms of miR-4262 on the regulation of cell proliferation and the cell cycle, the effects of miR-4262 mimic and inhibitor on cell proliferation- and cell cycle-related proteins such as PCNA, p21, cyclin D1, and CDK4 were detected by Western blot assay. The data showed that introduction of miR-4262 reduced the expressions of PCNA, cyclin D1, and CDK4 and enhanced the expression of p21. PCNA, a proliferation marker, is considered to be closely associated in the nuclear region for DNA synthesis31. Cyclin D1 (one of the cell cycle proteins) interacts with CDK4 to form the cyclin D1–CDK4 complex and to phosphorylate (p) retinoblastoma (Rb), which plays an important role in tumorigenesis. Previous studies have reported that the cyclin D1/CDK4/p-Rb signaling pathway is altered in many cancers32,33 and is an important regulator of the G1-to-S phase transition of the cell cycle. However, p21 belongs to a member of the CDK interacting protein/kinase inhibitory protein (Cip/Kip) family and blocks progression of the cell cycle through inhibition of the cyclin–CDK complex activity34. Collectively, these outcomes confirmed that miR-4262 affected cell proliferation by regulating PCNA, cyclin D1, CDK4, and p21. Additionally, EMT is one important molecular step in the process of distant metastasis, which is usually activated in many types of cancers including CC35,36. In EMT, genes such as E-cadherin, vimentin, N-cadherin, fibronectin, and β-catenin have been identified as the EMT markers37. After transfection with miR-4262 mimic and inhibitor, the changes of EMT markers in both HeLa and CaSKi cells were determined. The data demonstrated that upregulation of miR-4262 dramatically increased the epithelial marker E-cadherin and decreased the mesenchymal markers vimentin and β-catenin, which suggested that miR-4262 might inhibit the EMT process to regulate metastasis.
For further study, the data confirmed that Kaiso was a direct target of miR-4262 in CC cells. Several studies have demonstrated that increased Kaiso expression exists in many kinds of human cancers such as breast, prostate, non-small cell lung, and colorectal cancers21,22,24,38, indicating that Kaiso is considered to be a critical oncogene leading to the progression and metastasis of cancers. For example, Kaiso mediates the cyclin D1/cyclin E1/Rb1/E2F pathway, controlling passage through the G1 restriction point and accelerating cancer cell proliferation39. Kaiso also regulates miR-31 expression to accelerate cell migration and invasion of prostate cancer cells21. Bassey-Archibong et al. demonstrated that knockout of Kaiso can inhibit transforming growth factor-β (TGF-β) signaling and metastasis of triple-negative breast cancer cells40. In this study, restoration of Kaiso blocked the effects of miR-4262 mimic on the proliferation and EMT of CC cells.
In conclusion, the data demonstrated that the level of miR-4262 was dramatically decreased in CC tissues and cells. Overexpression of miR-4262 suppressed the proliferation and EMT of CC cells via directly targeting Kaiso. This novel miR-4262/Kaiso axis contributed to providing new insights into the mechanisms underlying the progression and metastasis of CC, and introduction of miR-4262 is considered to be a possible therapeutic strategy for treatment of CC in the future.
REFERENCES
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. [DOI] [PubMed] [Google Scholar]
- 2. Colombo N, Carinelli S, Colombo A, Marini C, Rollo D, Sessa C. Cervical cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2012;23(Suppl 7):i27–32. [DOI] [PubMed] [Google Scholar]
- 3. Castellsague X, Diaz M, de Sanjose S, Muñoz N, Herrero R, Franceschi S, Peeling RW, Ashley R, Smith JS, Snijders PJ, Meijer CJ, Bosch FX. Worldwide human papillomavirus etiology of cervical adenocarcinoma and its cofactors: Implications for screening and prevention. J Natl Cancer Inst. 2006;98(5):303–15. [DOI] [PubMed] [Google Scholar]
- 4. Ambros V. The functions of animal microRNAs. Nature 2004;431(7006):350–5. [DOI] [PubMed] [Google Scholar]
- 5. Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci USA 1999;96(8):4240–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell 2000;100(4):387–90. [DOI] [PubMed] [Google Scholar]
- 7. Sun Y, Zhang B, Cheng J, Wu Y, Xing F, Wang Y, Wang Q, Qiu J. MicroRNA-222 promotes the proliferation and migration of cervical cancer cells. Clin Invest Med. 2014;37(3):1–6. [DOI] [PubMed] [Google Scholar]
- 8. Shen L, Chen XD, Zhang YH. MicroRNA-128 promotes proliferation in osteosarcoma cells by downregulating PTEN. Tumour Biol. 2014;35(3):2069–74. [DOI] [PubMed] [Google Scholar]
- 9. Dang H, Zheng P, Liu Y, Wu X, Wu X. MicroRNA-543 acts as a prognostic marker and promotes the cell proliferation in cervical cancer by BRCA1-interacting protein 1. Tumour Biol. 2017;39(2):1–8. [DOI] [PubMed] [Google Scholar]
- 10. He Z, Xu H, Meng Y, Kuang Y. miR-944 acts as a prognostic marker and promotes the tumor progression in endometrial cancer. Biomed Pharmacother. 2017;88:902–10. [DOI] [PubMed] [Google Scholar]
- 11. Xia YF, Pei GH, Wang N, Che YC, Yu FS, Yin FF, Liu HX, Luo B, Wang YK. miR-3156-3p is downregulated in HPV-positive cervical cancer and performs as a tumor-suppressive miRNA. Virol J. 2017;14(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Su Z, Yang H, Zhao M, Wang Y, Deng G, Chen R. MicroRNA-92a promotes cell proliferation in cervical cancer via inhibiting p21 expression and promoting cell cycle progression. Oncol Res. 2017;25(1):137–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu D, Liu S, Zhang L, Song L. MiR-211 inhibits invasion and epithelial-to-mesenchymal transition (EMT) of cervical cancer cells via targeting MUC4. Biochem Biophys Res Commun. 2017;485(2):556–62. [DOI] [PubMed] [Google Scholar]
- 14. Fan JY, Fan YJ, Wang XL, Xie H, Gao HJ, Zhang Y, Liu M, Tang H. miR-429 is involved in regulation of NF-κB activity by targeting IKKβ and suppresses oncogenic activity in cervical cancer cells. FEBS Lett. 2017;591(1):118–28. [DOI] [PubMed] [Google Scholar]
- 15. Zhang D, Li Z, Zhang Y, Tu C, Huo J, Liu Y. miR-4262 promotes the proliferation of human cutaneous malignant melanoma cells through KLF6-mediated EGFR inactivation and p21 upregulation. Oncol Rep. 2016;36(6):3657–63. [DOI] [PubMed] [Google Scholar]
- 16. Wang K, Ren Y, Liu Y, Zhang J, He JJ. miR-4262 promotes proliferation and invasion of human breast cancer cells through directly targeting KLF6 and KLF15. Oncol Res. 2017;25(2):277–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu S, Wu J, Gao Y, Han G, Ding W, Huang X. MicroRNA-4262 activates the NF-κB and enhances the proliferation of hepatocellular carcinoma cells. Int J Biol Macromol. 2016;86:43–9. [DOI] [PubMed] [Google Scholar]
- 18. Song K, Liu N, Yang Y, Qiu X. Regulation of osteosarcoma cell invasion through osteopontin modification by miR-4262. Tumour Biol. 2016;37(5):6493–9. [DOI] [PubMed] [Google Scholar]
- 19. Collins T, Stone JR, Williams AJ. All in the family: The BTB/POZ, KRAB, and SCAN domains. Mol Cell Biol. 2001;21(11):3609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Daniel JM, Reynolds AB. The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol Cell Biol. 1999;19(5):3614–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang H, Liu W, Black S, Turner O, Daniel JM, Dean-Colomb W, He QP, Davis M, Yates C. Kaiso, a transcriptional repressor, promotes cell migration and invasion of prostate cancer cells through regulation of miR-31 expression. Oncotarget 2016;7(5):5677–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bassey-Archibong BI, Rayner LG, Hercules SM, Aarts CW, Dvorkin-Gheva A, Bramson JL, Hassell JA, Daniel JM. Kaiso depletion attenuates the growth and survival of triple negative breast cancer cells. Cell Death Dis. 2017;8(3):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones J, Wang H, Karanam B, Theodore S, Dean-Colomb W, Welch DR, Grizzle W, Yates C. Nuclear localization of Kaiso promotes the poorly differentiated phenotype and EMT in infiltrating ductal carcinomas. Clin Exp Metastasis 2014;31(5):497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dai SD, Wang Y, Miao Y, Zhao Y, Zhang Y, Jiang GY, Zhang PX, Yang ZQ, Wang EH. Cytoplasmic Kaiso is associated with poor prognosis in non-small cell lung cancer. BMC Cancer 2009;9(178):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dai SD, Wang Y, Zhang JY, Zhang D, Zhang PX, Jiang GY, Han Y, Zhang S, Cui QZ, Wang EH. Upregulation of δ-catenin is associated with poor prognosis and enhances transcriptional activity through Kaiso in non-small-cell lung cancer. Cancer Sci. 2011;102(1):95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Ann Rev Med. 2009;60:167–79. [DOI] [PubMed] [Google Scholar]
- 27. Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer 2006;6(4):259–69. [DOI] [PubMed] [Google Scholar]
- 28. Call JA, Eckhardt SG, Camidge DR. Targeted manipulation of apoptosis in cancer treatment. Lancet Oncol. 2008;9(10):1002–11. [DOI] [PubMed] [Google Scholar]
- 29. Molinari M. Cell cycle checkpoints and their inactivation in human cancer. Cell Prolif. 2000;33(5):261–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakanishi M, Shimada M, Niida H. Genetic instability in cancer cells by impaired cell cycle checkpoints. Cancer Sci. 2006;97(10):984–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sidhu GS, Singh AK, Banaudha KK, Gaddipati JP, Patnaik GK, Maheshwari RK. Arnebin-1 accelerates normal and hydrocortisone-induced impaired wound healing. J Invest Dermatol. 1999;113(5):773–81. [DOI] [PubMed] [Google Scholar]
- 32. Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10(8):789–99. [DOI] [PubMed] [Google Scholar]
- 33. Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10(7):699–703. [DOI] [PubMed] [Google Scholar]
- 34. Donjerkovic D, Scott DW. Regulation of the G1 phase of the mammalian cell cycle. Cell Res. 2000;10(1):1–16. [DOI] [PubMed] [Google Scholar]
- 35. Tian T, Li X, Hua Z, Ma J, Wu X, Liu Z, Chen H, Cui Z. S100A7 promotes the migration, invasion and metastasis of human cervical cancer cells through epithelial-mesenchymal transition. Oncotarget 2017;8(15):24964–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang WN, Li W, Wang XL, Hu Z, Zhu D, Ding WC, Liu D, Li KZ, Ma D, Wang H. CLDN1 expression in cervical cancer cells is related to tumor invasion and metastasis. Oncotarget 2016;7(52):87449–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pierre CC, Longo J, Mavor M, Milosavljevic SB, Chaudhary R, Gilbreath E, Yates C, Daniel JM. Kaiso overexpression promotes intestinal inflammation and potentiates intestinal tumorigenesis in Apc (Min/+) mice. Biochim Biophys Acta 2015;1852(9):1846–55. [DOI] [PubMed] [Google Scholar]
- 39. Pozner A, Terooatea TW, Buck-Koehntop BA. Cell-specific Kaiso (ZBTB33) regulation of cell cycle through cyclin D1 and cyclin E1. J Biol Chem. 2016;291(47):24538–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bassey-Archibong BI, Kwiecien JM, Milosavljevic SB, Hallett RM, Rayner LG, Erb MJ, Crawford-Brown CJ, Stephenson KB, Bédard PA, Hassell JA, Daniel JM. Kaiso depletion attenuates transforming growth factor-β signaling and metastatic activity of triple-negative breast cancer cells. Oncogenesis 2016;5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]