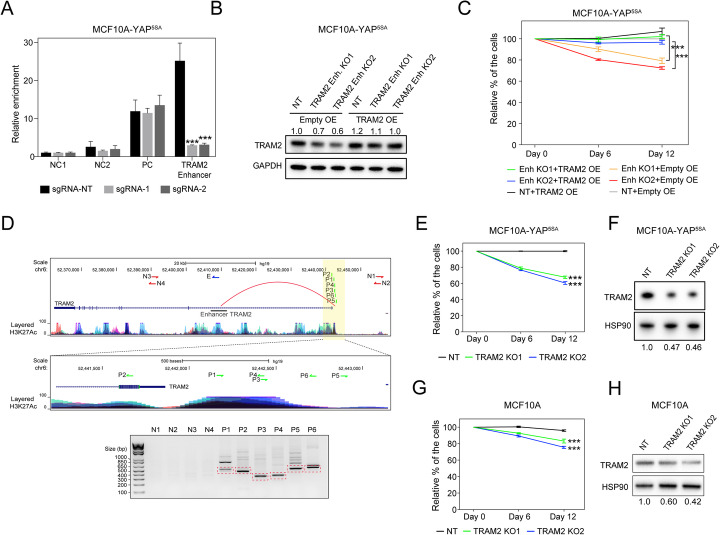
Fig. 3.
TRAM2 is a functional component of the YAP pathway. a ChIP-qPCR assays were used to quantify YAP-binding to EnhancerTRAM2 (chr6:52409174-52409286) in MCF10A-YAP5SA cells transduced with two different sgRNAs (1 and 2) targeting the TEAD4 motif within EnhancerTRAM2. A non-targeting sgRNA (sgRNA-NT) was used as control. Relative YAP binding was calculated and normalized to NC1 and input (NC1, negative control region1; NC2, negative control region2; PC, positive control region). Error bars indicate SD from three biological replicates. ***P < 0.005, two-tailed Student’s t test. b Western blot analysis with anti-TRAM2 antibody using lysates obtained from MCF10A-YAP5SA cells transduced with the indicated lentiviral vectors. NT, non-targeting sgRNA control. Anti-GAPDH antibody served as loading control. c Competitive proliferation of MCF10A-YAP5SA cells transduced with the indicated lentiviral vectors. Values on days 6 and 12 were normalized to day 0. Error bars indicate SD from three biological replicates. ***P < 0.005, two-tailed Student’s t test. NT, non-targeting sgRNA control; TRAM2 overexpression (TRAM2 OE), and control vector (Empty OE). d 3C chromatin capture experiments were used to assess the interaction between EnhancerTRAM2 and the promoter of TRAM2. Genome browser representation of the location for each primer used in the 3C analysis. A constant primer E (blue arrow) was used to amplify EnhancerTRAM2. Control regions (N1–N4) were amplified with primers indicated in red arrows, and the TRAM2 promoter region was amplified with primers indicated in green arrows (P1–P6). An agarose gel image of the PCR products with the expected sizes (red frame). Sanger sequencing results from the indicated PCR products are shown in Fig. S5A. e Competitive proliferation assay, as described in c, of TRAM2-KOs in MCF10A-YAP5SA cells. Values on day 6 (T = 6) and day 12 (T = 12) are normalized to day 0 (T = 0). Error bars indicate SD from three biological replicates. ***P < 0.005, two-tailed Student’s t test. f The cell populations used in e were subjected to a western blot analysis using anti-TRAM2 and control anti-HSP90 antibodies. g Competitive proliferation assay, as described in c, of TRAM2-KOs in MCF10A cells. Error bars indicate SD from three biological replicates. ***P < 0.005, two-tailed Student’s t test. h The cell populations used in g were subjected to a western blot analysis using anti-TRAM2 and control anti-HSP90 antibodies