FIG 3.
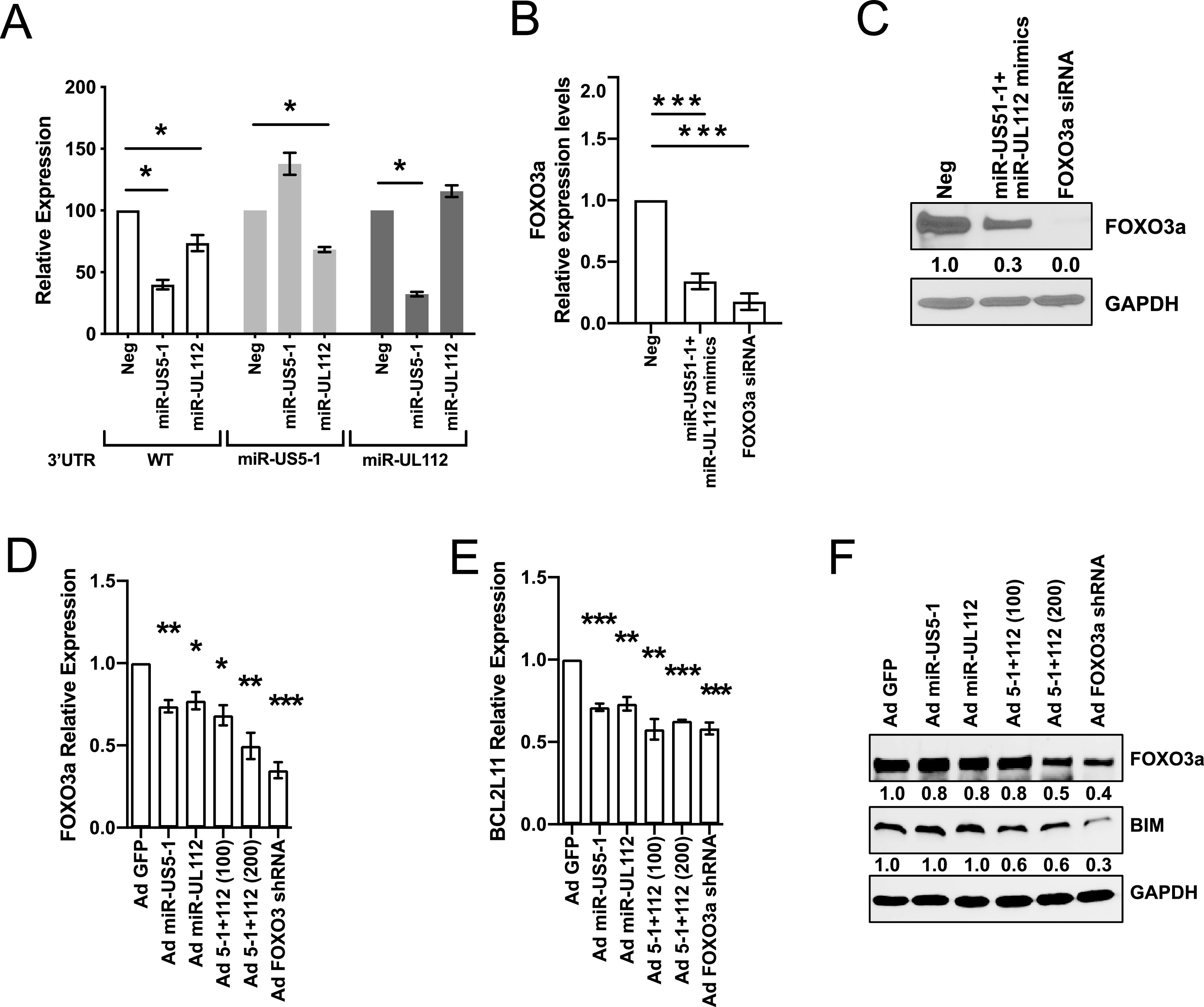
HCMV miR-US5-1 and miR-UL112-3p target FOXO3a protein for downregulation. (A) The wild-type (WT) FOXO3a 3′ UTR or 3′ UTRs containing mutations in the miR-US5-1 or miR-UL112 binding sites were cloned into the pSiCheck2 dual luciferase vector. HEK293T cells were transfected with the pSiCheck2 vector along with negative-control miRNA or miR-US5-1 or miR-UL112-3p mimic. Twenty-four hours later, cells were lysed and luciferase was measured. Values are means ± standard errors of the means (SEM) (error bars) for three independent experiments. Statistical significance was determined by unpaired Student’s t test (*, P < 0.05; **, P < 0.005; ***, P < 0.0005). (B) Normal human dermal fibroblasts (NHDF) were transfected with negative-control miRNA and miR-US5-1 and miR-UL112-3p mimics or a FOXO3a small interfering RNA (siRNA) for 48 h. RNA was isolated using TRIzol, and qRT-PCR for FOXO3a was performed. Values are means ± standard errors of the means (SEM) (error bars) for three independent experiments. Statistical significance was determined by unpaired Student’s t test (*, P < 0.05; **, P < 0.005; ***, P < 0.0005). (C) NHDF were transfected as in panel B, and protein was subjected to immunoblotting. Images were quantified using ImageJ and the ratio of FOXO3a to GAPDH protein, normalized to negative (Neg) control. (D and E) NHDF were serum starved and then transduced with the different adenoviral vectors for 48 h at an MOI of 200 or 100 when in combination. RNA was isolated using TRIzol, and qRT-PCR for FOXO3a (D) or BCL2L11 (E) was performed. Values are means ± standard errors of the means (SEM) (error bars) for three independent experiments. Statistical significance was determined by unpaired Student’s t test (*, P < 0.05; **, P < 0.005; ***, P < 0.0005). (F) NHDF were treated as described in panel D, and protein was subjected to immunoblotting. Images were quantified using ImageJ and the ratio of FOXO3a and BIM to GAPDH protein, normalized to Ad GFP control.
