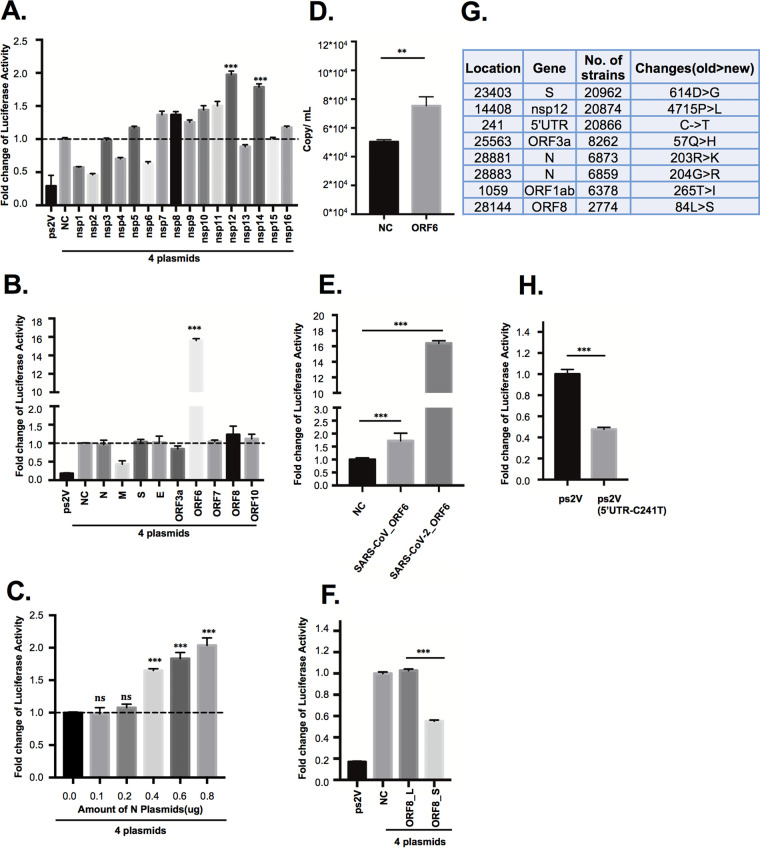
FIG 3.
Analysis of SARS-CoV-2 protein function based on the SARS-CoV-2 replicon system. (A, B, E, F, and H) ps2V (0.1 μg), ps2AN (0.05 μg), ps2AC (0.4 μg), ps2B (0.4 μg), and the respective mentioned plasmids (0.2 μg) were cotransfected into 293T cells (cell density: 6.5 × 104/cm2) seeded in a 12-well plate. The luciferase activity was detected by the luciferase reporter assay system after 48 to 54 h of transfection. (C) The gradient amounts of N-HA-expressing plasmid (consistency of plasmid amount of each group has been ensured by adding empty vector) were cotransfected into 293T cells with the combination of ps2V (0.1 μg), ps2AN (0.05 μg), ps2AC (0.4 μg), and ps2B (0.4 μg) plasmids. The luciferase activity was detected by the luciferase reporter assay system. (D) The authentic SARS-CoV-2 infection assay was performed according to Materials and Methods. At 48 h postinfection, virus RNA was isolated and RT-qPCR was performed to detect virus RNA copies. (G) The top 8 genome variations with high frequency based on the genome variation information obtained from the high-quality genome sequences of SARS-COV-2 collected from the NCBI and GISAID databases up to the date of 1 July 2020. The strain Wuhan-Hu-1 was retrieved from NCBI (NC_045512.2) as reference for variation annotation. The virus number with the variation is shown in the third column. All experimental data were analyzed using GraphPad Prism. All data are representative of at least three experiments.