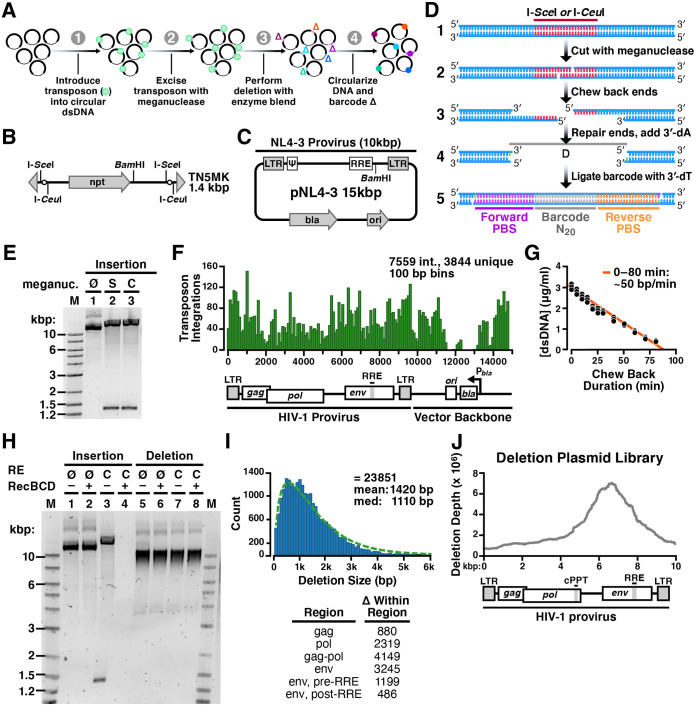
FIG 1.
Method to generate a random deletion library in HIV-1. (A) Overview schematic of method to create a barcoded random deletion library. (1) Transposon cassettes harboring unique restriction sites are inserted into plasmids via in vitro transposition. (2) Transposons are excised to linearize the insertion library with a meganuclease. (3) Deletions are performed by chewback from both DNA termini by simultaneous treatment with enzyme blend. Mean deletion size is modulated by adjusting duration of chewback. (4) The chewed termini are end repaired, dA tailed, and then joined by ligation to a T-tailed 60-bp unique barcode cassette. (B) Schematic of the “TN5MK” synthetic meganuclease transposon cassette used in library construction. TN5MK is composed of an antibiotic resistance gene, neomycin phosphotransferase I (npt), flanked by meganuclease restriction sites for I-SceI and I-CeuI and Tn5 mosaic ends (gray triangles) at the termini. The transposon cassette also contains a unique internal BamHI recognition site. (C) The HIV-1 molecular clone pNL4-3 is a 14,825-bp plasmid harboring the 9,709-bp NL4-3 provirus (HIV-1 subtype B). NL4-3 is a chimera of two viruses (NY5 and lymphadenopathy associated virus [LAV]). (D) Library insertion, excision, and barcoding details. (1) Circular DNA is linearized by digestion with a meganuclease (I-SceI or I-CeuI), which cleaves at recognition sites encoded on the inserted transposon. (2) This creates linear DNA with 4-base 3′ overhangs. Deletions are created by bidirectional chewback. (3) Treatment with two exonucleases (T4 and RecJf) creates a population of truncated deletion mutants with ragged ends. (4) Ragged DNA ends are blunted and then prepared for barcode cassette ligation by 5′ dephosphorylation and addition of a single 3′ dA. (5) Deletion mutants are religated in the presence of a barcode cassette with single 3′ dT overhangs and 5′ phosphoryl groups to create barcoded circular DNAs with 2 nicks separated by 60 bp. Barcodes are constructed with two primer-binding sites (PBS) on either side of a unique 20-bp sequence (barcode N20). (E) Insertion libraries following I-SceI (S) or I-CeuI (C) digestion. Digestion of pNL4-3 insertion library shows excisions of the TN5MK transposon (1.4 kb) and upward shift of the supercoiled library versus the undigested library. Lane M, 2-log DNA ladder; 1, undigested insertion library; 2, I-SceI digested insertion library; 3, I-CeuI digested insertion library. (F) Location of TN5MK insertions for a subset of 7,559 transposon integrations (3,844 were unique). (G) Determination of enzymatic chewback rate for deletion size. The chewback rate was determined by treating a 4-kb fragment of linear dsDNA with RecJf and T4 exonucleases in the presence of SSB and no dNTPs for increasing amounts of time and then halting enzymatic activity. Reactions were performed in triplicates. DNA concentrations were established by quantifying the fluorescence of PicoGreen in a plate reader in comparison to that of a dsDNA standard of known concentration. (H) Validation of deletion library. The pNL4-3 insertion library and pNL4-3 deletion library were either not digested (∅) or cut with I-CeuI (C) and then subjected to binary treatment with RecBCD, which digests linear DNA to completion. Lanes 1 to 4 are the pNL4-3 insertion library, and lanes 5 to 8 are the pNL4-3 deletion library. (I) pNL4-3 is composed of 23,851 tagged mutants with a range of deletion sizes. The right-skewed (i.e., right-tailed) histogram of deletion sizes in pNL4-3, with bins of 100 bp (shown in blue), is well-fit by a gamma distribution (green dashed line). (Inset) Number of deletions detected within each region of the HIV genome. (J) Deletion depth profile over the full HIV-1 genome. Calculation of the deletion depth profile of the pNL4-3 genome indicates that each base is covered by hundreds to thousands of deletion mutants. Two regions where deletions are not tolerated in the plasmid backbone are ori, the origin of replication, and bla, β-lactamase, the resistance marker.