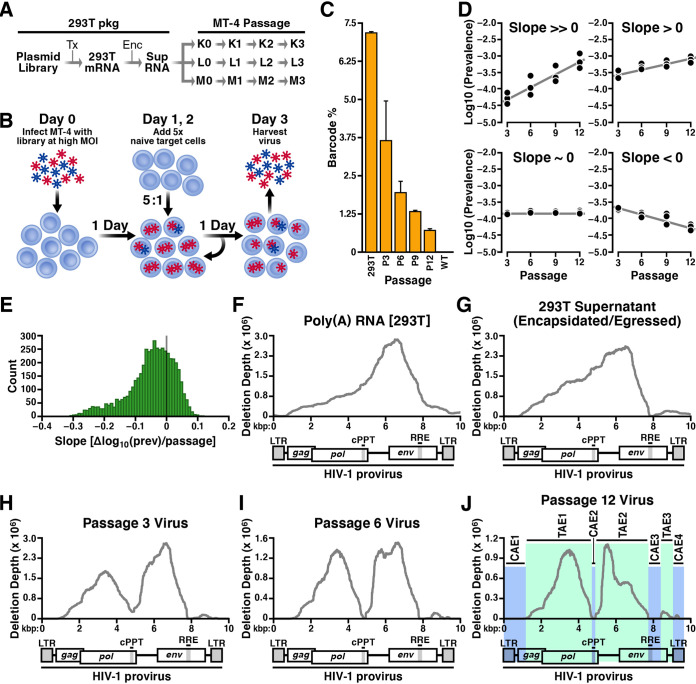
FIG 2.
Genetic screen of random deletion library to map viral cis and trans elements. (A) Block design of high-MOI passage. Wild-type NL4-3 and deletion library plasmids cotransfected 293T cells. Virus-containing supernatant-infected MT-4 cells in triplicates (K, L, M) at high MOI. Supernatant was collected and transferred onto new cells at the end of every week, for 4 weeks. At the same time, flasks with only NL4-3 wild-type virus were passaged identically. (B) Passage details of high-MOI screen. MT-4 cells (blue double discs) are infected at high MOI with a pool of virus (HIV-1) containing both wild-type (red stars) and deletion mutants (blue stars). At days 1 and 2, additional naive MT-4 cells were added and the culture volume expanded. On day 3, cell-free supernatant was harvested and virus purified by ultracentrifugation for transfer or analysis. There were 3 total rounds of HIV replication from day 0 to day 3. (C) Detection and quantification of barcode cassettes by RT-qPCR. Genomic percentages of barcoded mutants to total HIV genomes in transfection (293T), each stage of high-MOI passage (P3 to P12), and a wild-type HIV control (WT). RT-qPCR data were normalized to an MS2 RNA spike-in. Error bars are standard deviations from averaging each flask (K, L, and M) per passage. (D) Representative deletion variant trajectories during high-MOI passage. The slope in prevalence versus passage number was determined by linear regression. Data points correspond to the triplicate flasks (K, L, and M) at each passage. Prevalence is in reference to the total barcode cassette pool (tagged mutants). (E) Distribution of fitness in deletion variants that are not extinct by passage 12. The vertical line marks a slope of 0 for reference. (F) The deletion depth profile of poly(A) RNA from transfected 293T cells, representing mutants able to be transcribed. (G) The deletion depth profile built from the virus-containing supernatant of transfected 293T representing mutants able to be transcribed, encapsulated, and egressed. (H) Deletion depth profile from virus-containing supernatant after 3 passages. (I) Deletion depth profile after 6 passages. (J) A model of HIV-1 cis- and trans-acting elements after 12 passages. The HIV-1 genome is composed of 4 cis-acting elements, CAE1 to CAE4 (highlighted in blue), and 3 trans-acting elements, TAE1 to TAE3 (highlighted in green).