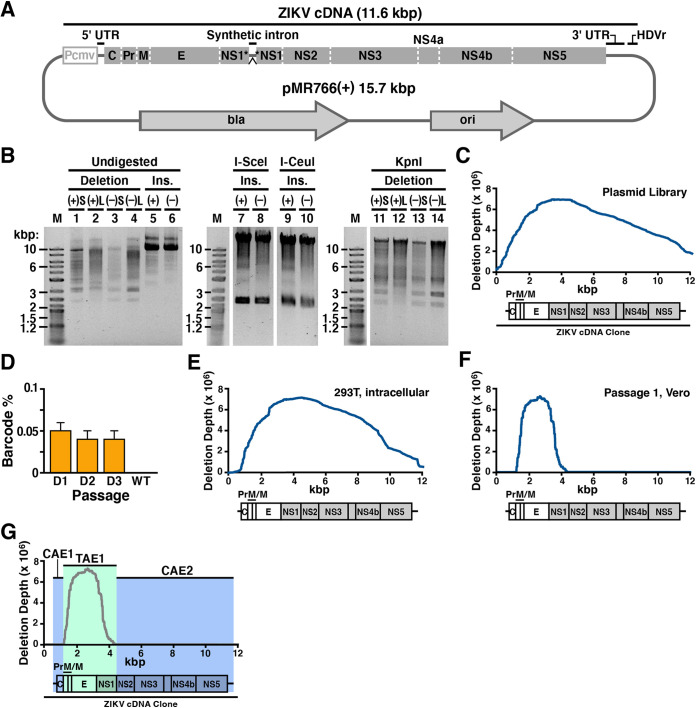
FIG 4.
Application of RanDeL-seq to map Zika virus (ZIKV) cis elements. (A) pMR766(+), a Zika virus molecular clone. The MR766 Zika virus genome is encoded as a cDNA driven by the CMV IE2 promoter. At the 3′ end of the genome, a self-cleaving hepatitis delta virus ribozyme allows for creation of an authentic 3′ end posttranscription. An intron sequence is present within NS1 to allow maintenance in bacteria but is spliced out during transcription in host cells. (B) Restriction enzyme characterization of completed ZIKV deletion libraries compared to insertion libraries (“Ins.”). (+) and (−) designate the template ZIKV plasmid. “S” and “L” designate the chewback length for deletion libraries. Undigested completed deletion libraries (lanes 1 to 4) were run next to undigested insertion libraries (lanes 5 and 6). Insertion libraries (lanes 7 to 10) treated with I-SceI or I-CeuI to excise transposon (∼1.4 kb). Deletion libraries linearized by unique ZIKV cutter KpnI (lanes 11 to 14). (C) Deletion depth profile of the pMR766(+)L library. The ZIKV genome is well represented in the pMR766(+)L library, with some bias. Each base of the ZIKV genome is covered by several hundred different deletion mutants. (D) Detection and quantification of ZIKV barcode cassettes by RT-qPCR. Genomic percentages of barcoded mutants to total ZIKV genomes at each day in passage 1 of the high-MOI screen and a wild-type ZIKV control (WT). RT-qPCR data were normalized to an MS2 RNA spike-in. (E) Deletion depth profile of intracellular RNA of 293T cotransfected with the wild-type ZIKV plasmid and the pooled deletion libraries. (F) Deletion depth profile of pMR766(+)L after passage 1. Only deletions in Pr to NS1 can be trans-complemented by wild-type ZIKV. (G) Final map of ZIKV cis- and trans-acting elements after passage 2. The two cis-acting regions are highlighted in blue and do not tolerate deletion (i.e., must be present for efficient transmission to occur). The trans-acting region is highlighted in green and can be complemented in trans (i.e., if deleted, transmission occurs by complementation from wild-type virus).