Abstract
Intravascular ultrasound (IVUS) imaging is widely used for diagnostic imaging in interventional cardiology. The detection and quantification of atherosclerosis from acquired images is typically performed manually by medical experts or by virtual histology IVUS (VH-IVUS) software. VH-IVUS analyzes backscattered radio frequency (RF) signals to provide a color-coded tissue map, and is the method of choice for assessing atherosclerotic plaque in situ. However, a significant amount of tissue cannot be analyzed in reasonable time because the method can be applied just once per cardiac cycle. Furthermore, only hardware and software compatible with RF signal acquisition and processing may be used. We present an image-based tissue characterization method that can be applied to entire acquisition sequences post hoc for the assessment of diseased vessels. The pixel-based method utilizes domain knowledge of arterial pathology and physiology, and leverages technological advances of convolutional neural networks to segment diseased vessel walls into the same tissue classes as virtual histology using only grayscale IVUS images. The method was trained and tested on patches extracted from VH-IVUS images acquired from several patients, and achieved overall accuracy of 93.5% for all segmented tissue. Imposing physically-relevant spatial constraints driven by domain knowledge was key to achieving such strong performance. This enriched approach offers capabilities akin to VH-IVUS without the constraints of RF signals or limited once-per-cycle analysis, offering superior potential information acquisition speed, reduced hardware and software requirements, and more widespread applicability. Such an approach may well yield promise for future clinical and research applications.
Keywords: Atherosclerosis, IVUS, Convolutional Neural Networks, Deep Learning, Plaque Characterization
I. Introduction
Atherosclerosis is an inflammatory disease which scleroses and obstructs flow through arterial blood vessels [1], [2]. Atherosclerotic plaques composed of lipids, inflammatory cells, and calcium deposits form in the vessel wall and ultimately impinge on the lumen, reducing distal perfusion. Tissue insufficiency that follows causes diseases that are the leading cause of morbidity and mortality globally [3].
A primary step in diagnosing and treating atherosclerosis is imaging the arterial vessel wall. Though several techniques can visualize the lumen border and roughly ascertain the constitution of the arterial wall, intravascular imaging is the current method of choice in interventional cardiology [4]-[6]. Intravascular ultrasound (IVUS) is an invasive technique which provides two-dimensional (2D) tomographic views of the coronary lumen and vessel wall, allowing comprehensive visualization of any plaque. Generated images can provide reliable geometric measurements and estimates of plaque composition [7]. A well-trained expert can manually determine the dimensions of the lumen and media-adventitia border. Together these delineate the limits of the arterial wall and primary region of interest (ROI), as well as four different plaque constituent types: dense calcium (DC), necrotic core (NC), fibrotic tissue (FT), and fibro-fatty tissue (FFT) [8], [9]. DC is composed of compact calcium crystals, while NC consists of high levels of lipids with many necrotic cells. While both FT and FFT include collagen fibers, the former is mainly bundles of fibers [10], and the latter loosely packed fibers with lipid accumulations [11]. Due to their varying composition, each plaque type has unique echoreflectivity characteristics and consequently differentiable appearance within an IVUS image.
Manual ROI and tissue detection has been used since the introduction of IVUS. However, acquisition sequences can contain several thousand individual frames (images) [7], so manual processing is time-consuming and laborious. It is also subject to high inter- and intra-observer variability [12]. Moreover, discrimination of FT from FFT is limited, since the two plaques share similar characteristics. These limitations led to the development of automated ROI detection algorithms [13]-[18] and methods to segment tissue within the arterial wall [4].
Numerous plaque characterization methods using IVUS images have been reported in the literature. The majority of these methods is based on machine learning approaches. The first methodology was presented by Zhang et al. [19], who automatically extracted image texture features and classified pixels using a learned piecewise linear discrimination function. Since then, many have followed, using different feature sets and classification algorithms [20], [21]. Such methods follow the same general pattern: grayscale images are used as input and pixels are classified by a machine learning algorithm according to the pixels’ intensities and imaging characteristics (e.g. acoustic shadows) or a supplementary set of extracted texture and geometric features. The gold standard for those methods was human expert manual annotations, which limited the amount of available data and suffered from inter- and intra-observer variability; subsequent implementation of the methods in clinical practice was hindered in part because validation and training relied upon such manual annotations. Therefore, Taki et al. [22], [23] – followed by others [24]-[26] – proposed similar machine learning approaches trained and validated using the results of a commercially available software: virtual histology (VH) IVUS [11].
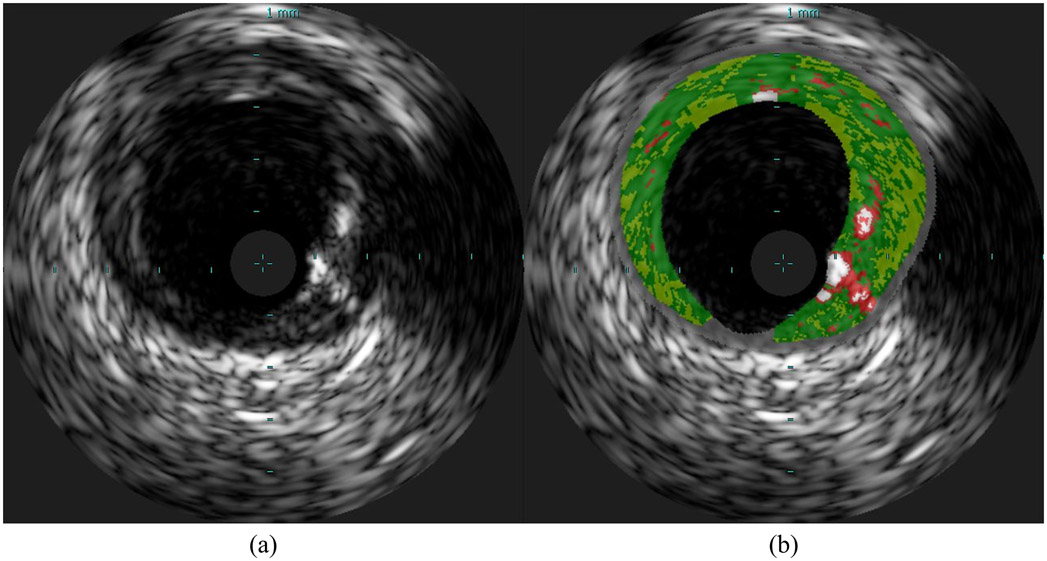
VH-IVUS was introduced to surmount the limitations of manual labeling of diseased vessels [11]. VH-IVUS offers a color-coded plaque characterization map, often overlaid on the corresponding grayscale image (Fig. 1). By processing the frequency spectrum of backscattered radiofrequency (RF) signal [27], rather than just the reflected signal amplitude, a more detailed assessment of the plaque can be generated with high accuracy confirmed through histology validation [8], [11], [28]-[30]. VH-IVUS can classify plaque into its four subtypes [11], and treats the non-pathological tissue and media – the concentric layer separating the disease-prone intima from the outer adventitia layer – as a separate combined class (M). The technology is the current gold standard for in vivo and in situ examination of coronary arteries [8], [11]. Although VH-IVUS provides relatively accurate plaque characterization, its main disadvantage is the fact that it requires acquisition of RF signal and proprietary software to process this signal. As a consequence, the plaque composition of grayscale IVUS frames acquired without the full RF signal (or without the proprietary software) cannot be characterized by this technique. Moreover, the RF signal is available only in the ECG-gated R-peak IVUS frames [31] – ~1 of every 30 frames – resulting in significant information loss and large segments of uncharacterized vessel. Thus, methods able to characterize the plaque in a similar manner as VH-IVUS using grayscale methods remain attractive and highly relevant.
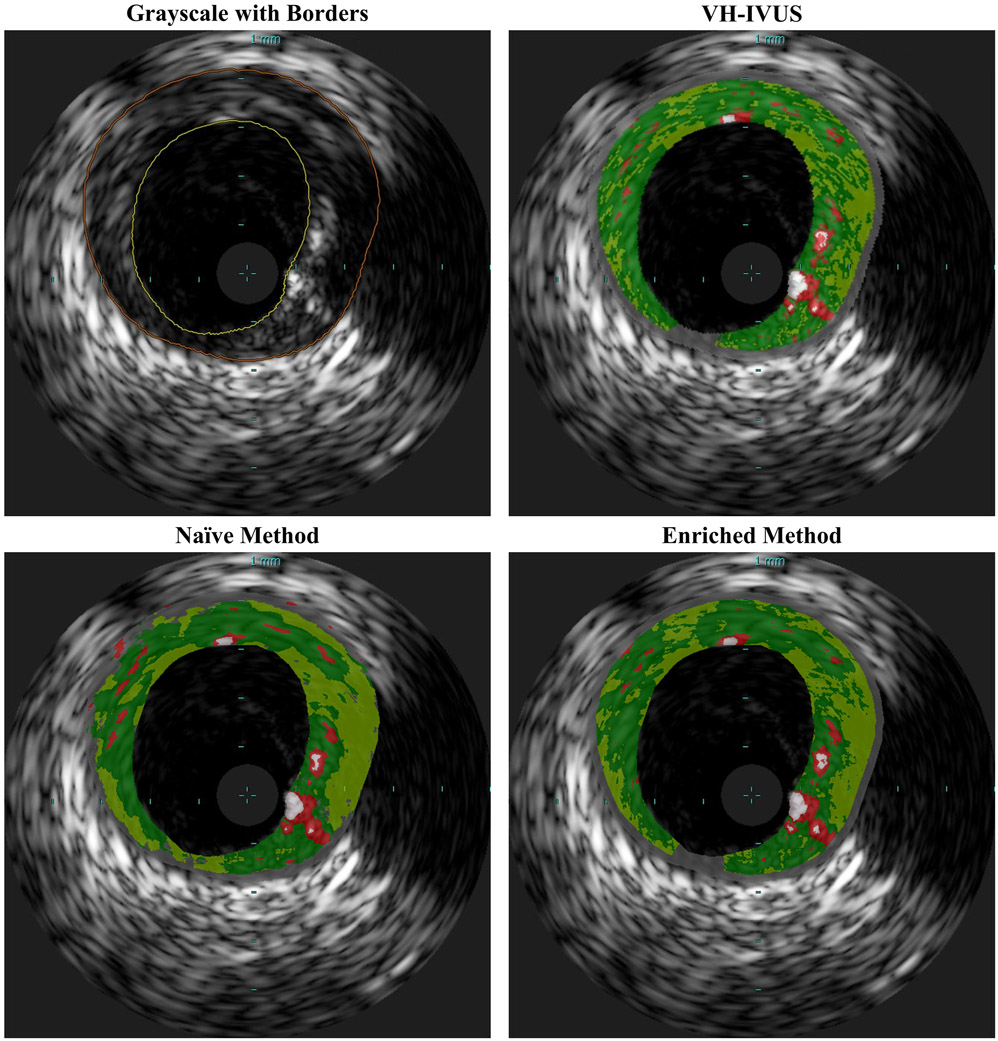
Fig. 1.
Sample VH-IVUS frame: (a) Grayscale IVUS image and (b) the same image overlaid with plaque types characterized by VH as dense calcium (DC; white), necrotic core (NC; red), fibrotic tissue (FT; green), fibro-fatty tissue (FFT; light green), and media or non-pathological tissue (M; gray).
Recent developments in deep learning and convolution neural networks (CNN) have made possible characterization tools in different imaging modalities which outperform methods deploying traditional machine learning or image processing [32]. Indeed, none of the existing IVUS plaque characterization methods, which require explicit feature set design, selection, and extraction through pre-processing, have achieved overall label assignment accuracy >90% [4] (Table III). To date, however, deep learning has been applied to IVUS only for delineating inner and outer boundaries of the arterial wall (i.e. ROI) [17], [18] and to select frames containing calcification [33]; no method has applied CNNs to grayscale IVUS imaging data to improve plaque characterization and generate information akin to VH-IVUS.
TABLE III.
Comparison of Current Methods & Previous Methods Reported in Literature
| Method | Tissue Classes a | Gold Standard | Validation Dataset | Reported Accuracy |
|---|---|---|---|---|
| Zhang et al. [19] | DC, NC, & FT | Expert annotations | 12 IVUS images | Overall: 89.9% |
| Brunenberg et al. [20] | DC, NC, & FT | Expert annotations | 30 IVUS images | Overall: 75.82% |
| Athanasiou et al. [21] | DC, NC, & FT | Experts annotations | 40 IVUS images | Overall: 86.17% |
| Taki et al. (2010) [22] | DC, NC, & FT-FFT | VH-IVUS | 500 VH-IVUS frames | DC: 79%, NC: 52%, & FT-FFT: 81% |
| Taki et al. (2013) [23] | DC, NC, & FT-FFT | VH-IVUS | 500 VH-IVUS frames | DC: 84.87%, NC: 80.57%, FT: 77.4%, & FFT: 63.47% |
| Athanasiou et al. [24] | DC, NC, FT, FFT, & M | VH-IVUS | 300 VH-IVUS frames | DC: 84.87%, NC: 80.57%, FT: 77.4%, & FFT: 63.47% |
| Hwang et al. [25] | DC, NC, FT, & FFT | VH-IVUS | 252 VH-IVUS frames | Overall: 82.8% |
| Kim et al. [26] | DC, NC, FT, & FFT | VH-IVUS | 252 VH-IVUS frames | Overall: 85.1% |
| Naïve Deep Learning Approach | DC, NC, FT, FFT, & M | VH-IVUS | 200 VH-IVUS frames | DC: 98.7%, NC: 87.3%, FT: 87.5%, FFT: 89.5%, & M: 75.9% |
| Domain Enriched Deep Learning Approach | DC, NC, FT, FFT, & M | VH-IVUS | 200 VH-IVUS frames | DC: 98.5%, NC: 88.6%, FT: 91.1%, FFT: 90.0%, & M: 99.4% |
| VH-IVUS (Validation by Nair et al. [27]) | DC, NC, FT, FFT, & M | Histology | 889 histological regions | DC: 96.7%, NC: 95.8%, FT: 93.5%, & FFT: 94.1% (M not reported) |
DC: Dense Calcium; NC: Necrotic Core; FT: Fibrous Tissue; FFT: Fibro-Fatty Tissue; M: Media/Non-Pathological
We present a novel CNN-based domain enriched method that classifies arterial tissue imaged through IVUS. The method detects the ROI using recently developed software [34], and then subdivides the ROI into pathological and non-pathological tissue based upon basic spatial and geometric constraints informed by physiology. Pathological areas of the ROI are partitioned into patches and fed through a CNN architecture. Corresponding VH-IVUS images serve as the comparative control. The proposed method offers several meaningful benefits stemming from its independence from the RF signal data, which increases the clinical utility and research applicability of the method. In particular, the method can be applied to grayscale IVUS data, including previously-acquired images that have not been characterized by the VH technique due to a lack of RF signal or proprietary software, or to intermediate frames of VH-IVUS acquisitions between ECG-gated frames, thereby increasing the effective rate at which meaningful information on plaque morphology can be attained and reducing procedure time.
II. Materials and Methods
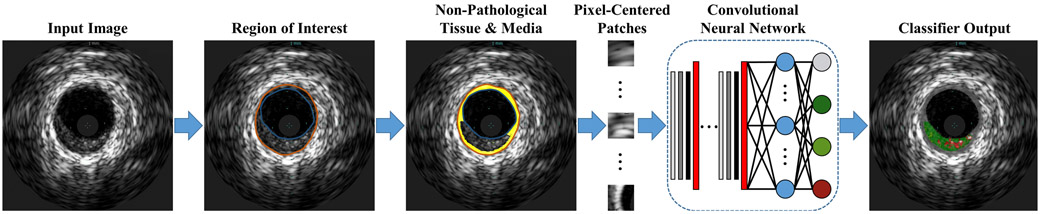
The proposed automated plaque characterization method consists of three steps (Fig. 2). The ROI is first detected, then pathological tissue is partitioned from the rest of the vessel wall (M) based upon domain knowledge of spatial constraints imposed by arterial physiology and pathology. This process imposes physically-relevant limits on the location and dimensions of this tissue class while also reducing the number of classes to be subsequently segmented by the CNN. In the final step, pixels of the ROI in the pathological area are classified into one of the four plaque types. To investigate the utility of leveraging domain enrichment, an equivalent “naïve” method was implemented where non-pathological tissue was not first segmented from the pathological tissue prior to CNN segmentation, but was instead segmented as a fifth class. The method was implemented in MATLAB (MathWorks, Natick, MA) using the Deep Learning Toolbox running on a NVIDIA TITAN Xp GPU (PG611) with 12 GB RAM.
Fig. 2.
Flowchart of the plaque characterization method enriched by domain knowledge. The naïve method does not segment the non-pathological tissue and media based upon vascular physiology and pathology constrains, but rather inputs the full ROI to the CNN, which must segment the image into all 5 classes.
A. Region of Interest
The region between the lumen border and the media-adventitia border where atherosclerotic plaques develop was denoted as the ROI. ROI segmentation is a prerequisite for subsequent methodological steps, though succeeding procedures are agnostic to ROI segmentation approach, method, or algorithm (of which there are a large and growing number). To detect the ROI in each frame, we here utilized a previously validated method [13] recently incorporated into a user-friendly software suite [34]. In brief, initial contours for the lumen and media-adventitia borders are estimated using basic image processing: the image is binarized using Otsu’s automatic thresholding algorithm [35], and the tentative borders are found by scanning radial projections for binary state transitions. The method subsequently refines the borders using active contour models [36]. Within each IVUS image I(i, j), the lumen border bl(θ) and media-adventitia border bma(θ) fully delineate the ROI (intima and media region) rim(irim,jrim).
B. Pathological Tissue Detection
The proposed method focuses on the evaluation of vessel wall morphology and the characterization of its phenotype, distinguishing not only plaque subtype but normal from pathological tissue. This concept has already been implemented in VH-IVUS, where each tissue type is highlighted as a specific color and the media portrayed in gray along the rim of the vessel wall (Fig. 1). Physical and dimensional limits were imposed herein, leveraging expert recommendations for interpreting intravascular images; intima was deemed normal if its thickness was <360 μm, and the media was assumed have nominal thickness of 250-350 μm [31], [37], [38]. Thus, the location and thickness of non-diseased and media tissue was defined such that wall regions thinner than threshold were not to be considered diseased or analyzed as such, and the media layer approximated by a band of constant thickness around the outer edge of the ROI. Though media thickness does vary somewhat, its range is largely negligible relative to that of the inner intima layer, and is furthermore at the horizon of VH-IVUS imaging resolution (100-200 μm) [7], [9], [31].
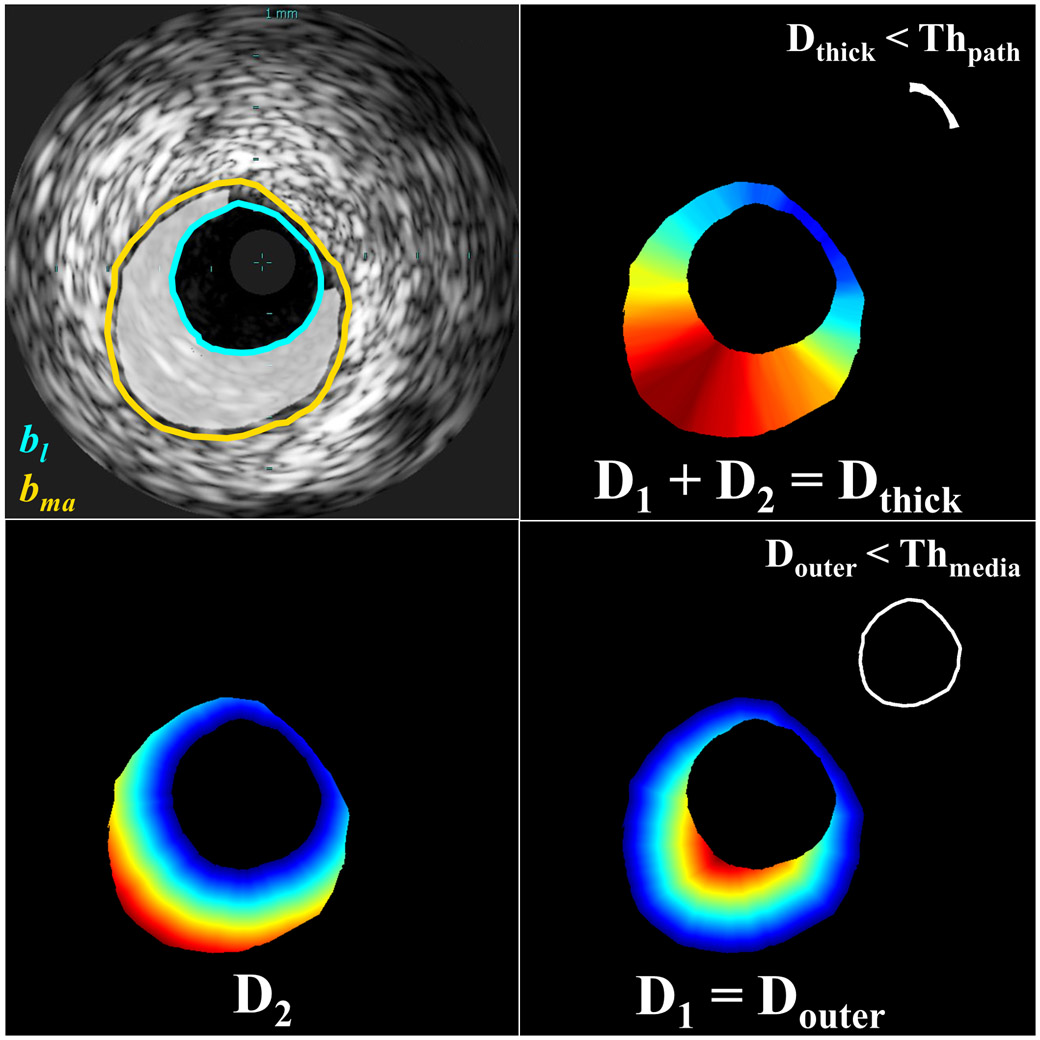
To determine the normal wall and the media layer locations and dimensions, two geometrical parameters were computed for each pixel in the ROI:
| (1) |
| (2) |
where D1 and D2 are the Euclidian distances of the pixel (irim,jrim) from the media-adventitia border bma and the lumen border bl, respectively (Fig. 3 and Fig. S1). Threshold values for Dthick and Douter were calculated to determine whether a pixel was in a section of sufficient thickness to be considered pathological or sufficiently close to the media-adventitia border to lie within the media. All Ntot VH-IVUS images and their ROI pixels that belong to the media or non-pathological class (M, gray color; ) were considered. The pathological thickness threshold was calculated as the maximum section thickness immediately adjacent to the lumen (bl):
| (3) |
Fig. 3.
Schematic presentation of the pathological tissue segmentation. Given borders of the lumen (bl) and media-adventitia (bma; top left), Euclidean distances from a pixel (rim ∈ ROI) to the lumen border (D2) and the media-adventitia border (D1) were calculated (bottom). Pixels within the ROI for which Douter < Thmedia and Dthick < Thpath correspond to media and non-pathological tissue, respectively (right, inset). Other pixels within the ROI correspond to pathological tissue (ROIpath; top left, highlighted). Color in distance maps indicates relative magnitude of values (blue: small, red: large).
The maximum media thickness threshold was calculated as the minimum thickness of sections in which pathological tissue is present (i.e. Dthick ≥ Thpath):
| (4) |
Thpath was 30 pixels, and Thmedia was 11 pixels. Pixels of the ROI were classified as pathological tissue (ROIpath) if Douter ≥ Thmedia and Dthick ≥ Thpath (Fig. 3).
This pathological tissue detection procedure is the primary mechanism by which domain knowledge enriched learning to address the image classification problem. Following this step, classification was only required for the four remaining tissue types. For the naïve method developed to assess the importance of this contribution, this step was not completed; instead, subsequent classification routines were taught to detect this tissue type directly from the image patch data.
C. Classification
For the domain enriched method, pixel-centered patches were created for remaining pixels of the ROI after segmenting the M class (rim ∈ ROIpath), then automatically classified into one of the four plaque types using a CNN. For the naïve method, patches were created for all pixels of the ROI (rim ∈ ROI) and sorted into one of the five tissue types by the classifier.
1). CNN Algorithm
CNNs are a class of deep neural networks [39] commonly applied to image classification because they can leverage spatial locality and translational invariance to dramatically reduce the number of weighted network connections requiring optimization (cf. fully-connected neural networks). Their architectiue can be described by multiple layers, which can be categorized as input, output, or hidden. The input layer here receives the 2D (grayscale) image patch, the hidden layers are formed by multiple functional layers in which the compound image featiues are calculated and strategically pooled, and the output layer is the classification result. Combined in series, such a CNN can be represented by a non-linear function, P(I; Θ) = pi,which maps an image of H × H size to a vector pi = (p1, p2, … , pc)T. The probability of I belonging to one of target classes i = {1, … , c} is represented by pi ∈ [0,1], and Θ = {Θ1, Θ2, … , ΘK} are the K parameters (weights and biases) used to map I to pi. CNN training is an optimization problem for a non-linear function with many degrees of freedom:
| (5) |
where is a loss function and Ntrain is the number of training images.
Here, we used multiclass cross-entropy loss (also known as negative log likelihood), the most popular choice for probabilistic classification problems:
| (6) |
This loss function measures the performance of the classifier P relative to the binary class label vector yi.
To reduce the training time for the CNN, the stochastic gradient descent (SGD) iterative method was used. This method approximates the dataset with a subset of samples randomly-drawn from the frill training dataset, called a mini-batch, and uses the gradient calculated for the mini-batch to update the model in each iteration. SGD is known to sometimes oscillate along the path of steepest descent (maximum gradient) towards the optimum, rather than directly along the path toward the optimum, since the gradient always points towards the opposite side of this optimum from the current position. A solution to this problem is the addition of a momentum term to the parameter update to reduce oscillations:
| (7) |
where λ is the iteration number, α > 0 is the learning rate, and the momentum term γ determines the contribution of the previous gradient step to the current iteration. Thus, the SGD algorithm selects a subset of the training set , evaluates the mean gradient of the loss function for this mini-batch, then updates the network parameters Θ. Each evaluation is an iteration, and at each iteration the loss function is minimized further. The full pass of the training process over the whole training set, in mini-batch increments, forms an epoch.
In training the network described herein, a stochastic gradient descent with momentum optimizer was implemented with a constant learning rate (α) of 0.03 and momentum value (γ) of 0.9. A mini-batch size of 3,000 patches was utilized over 50 epochs; data were shuffled after each epoch. Weight decay (L2 regularization) by a factor of 0.0001 was used to reduce overfitting. Weights were initialized with a Glorot initializer, which independently samples from a uniform distribution centered around zero; biases were initialized to zero.
2). CNN Architecture
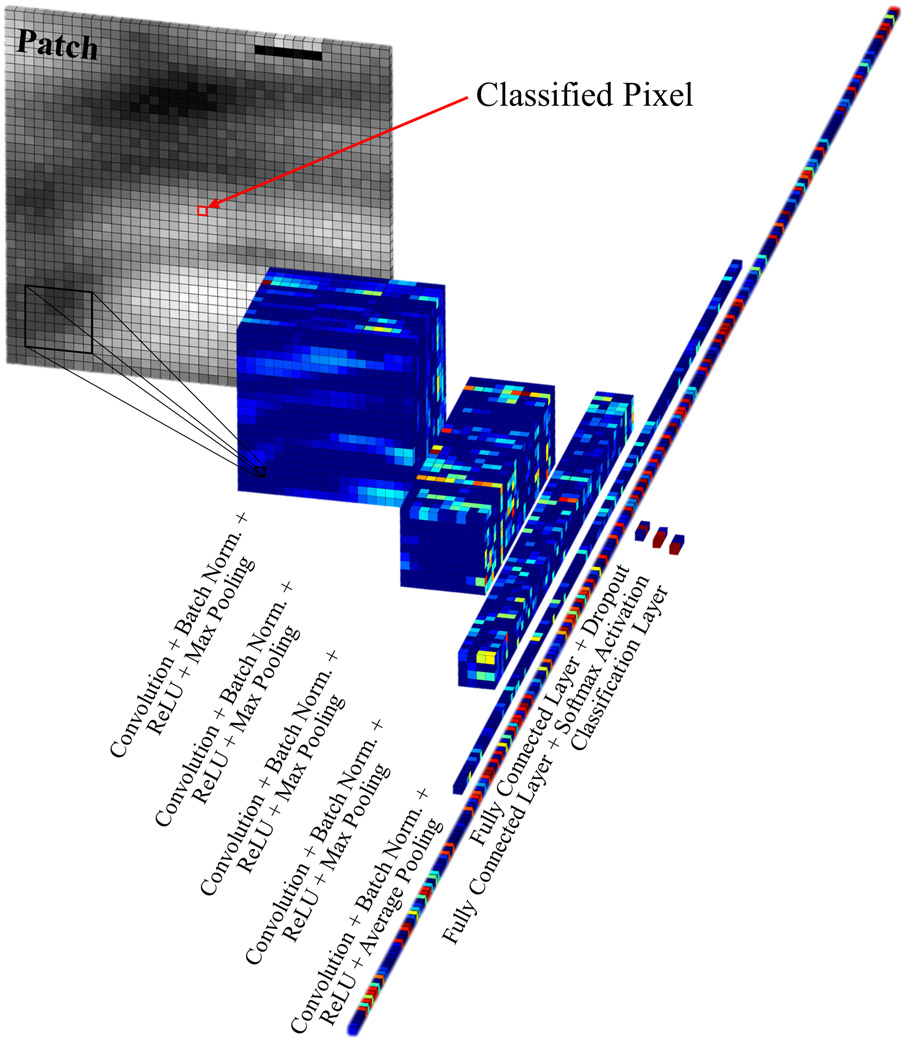
To classify the pixels corresponding to pathological tissue, a sequence of convolutions, activations, and pooling operations were executed. To achieve the best classification results, different patch sizes, numbers of input patch convolution sequences, filters, and filter sizes were tested. A patch size of 41×41 was determined to perform best through parameter sensitivity analysis (Fig. S6). The network found to perform best, and utilized in this work, is shown in Fig. 4 and Fig. S2 (Supplemental Materials).
Fig. 4.
Progressive data processing performed by the 26-layer CNN to classify pixels within the pathological region of interest. (See Fig. S2 in Supplemental Materials for a detailed schematic of the CNN architecture.)
III. Dataset
To train and test our plaque characterization algorithm, 553 VH-IVUS frames and the corresponding grayscale IVUS frames were acquired from eight patients. The data were acquired at 20 MHz using a 3.5 F electronic probe with synthetic aperture (Eagle Eye Gold Catheter, Philips Healthcare, Andover, MA), in accordance with clinical standards [7], [31]. From the dataset, 200 frames were withheld exclusively for testing while the remaining frames were sampled for training and validation. From this larger subset, equal numbers of 41-by-41 pixel patches (3.4×105) were randomly extracted for each of the five classes, and data augmentation was performed through reflection and rotation in 90° increments. From the withheld testing subset, 5×104 patches of each class were randomly selected from bulk regions of tissue for final testing and validation. Additional details on the dataset are available in the Supplemental Materials.
IV. Results
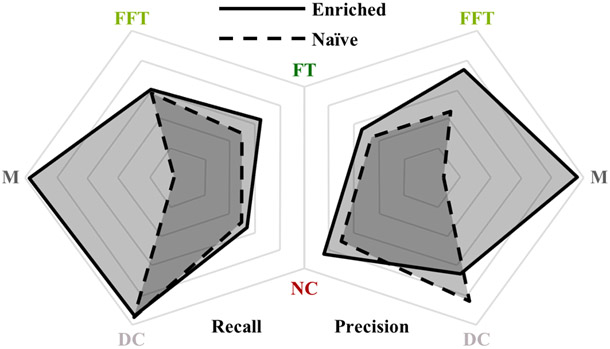
Image segmentation accurately replicating VH-IVUS classification was successfully achieved using only grayscale IVUS images, with the domain enriched method providing better results than the naïve one. Tables I and II provide the error (or confusion) matrices for the enriched and naïve methods, respectively, showing that the former achieved an overall accuracy of 93.5% and the latter 87.8%. Performance metrics by tissue class are summarized and compared in Fig. 5.
TABLE I.
Domain Enriched: Spatial Constraints + ROIpath Segmentation
| Target Class | |||||||
|---|---|---|---|---|---|---|---|
| DC | NC | FT | FFT | M | Precision | ||
| Output Class | DC | 49247 | 4664 | 0 | 0 | 3 | 91.3% |
| NC | 664 | 44310 | 1208 | 0 | 30 | 95.9% | |
| FT | 0 | 996 | 45564 | 4761 | 159 | 88.5% | |
| FFT | 0 | 0 | 3107 | 45015 | 84 | 93.4% | |
| M | 89 | 30 | 121 | 224 | 49724 | 99.1% | |
| Recall | 98.5% | 88.6% | 91.1% | 90.0% | 99.4% | 93.5% | |
TABLE II.
Naïve: Full-ROI Segmentation
| Target Class | |||||||
|---|---|---|---|---|---|---|---|
| DC | NC | FT | FFT | M | Precision | ||
| Output Class | DC | 49371 | 1840 | 1 | 0 | 123 | 96.2% |
| NC | 506 | 43652 | 978 | 0 | 2040 | 92.5% | |
| FT | 0 | 477 | 43747 | 2168 | 4222 | 86.4% | |
| FFT | 0 | 0 | 1408 | 44738 | 5686 | 86.3% | |
| M | 123 | 4031 | 3866 | 3094 | 37929 | 77.3% | |
| Recall | 98.7% | 87.3% | 87.5% | 89.5% | 75.9% | 87.8% | |
Fig. 5.
Comparison of recall (i.e. sensitivity) and precision (i.e. positive predictive value) achieved by the enriched and naïve methods (shown with solid and dashed borders, respectively). The enriched method demonstrates clear superiority, particularly, but not exclusively, in categorizing M class tissue. Axes range from 75% to 100% (linear scale from center to perimeter).
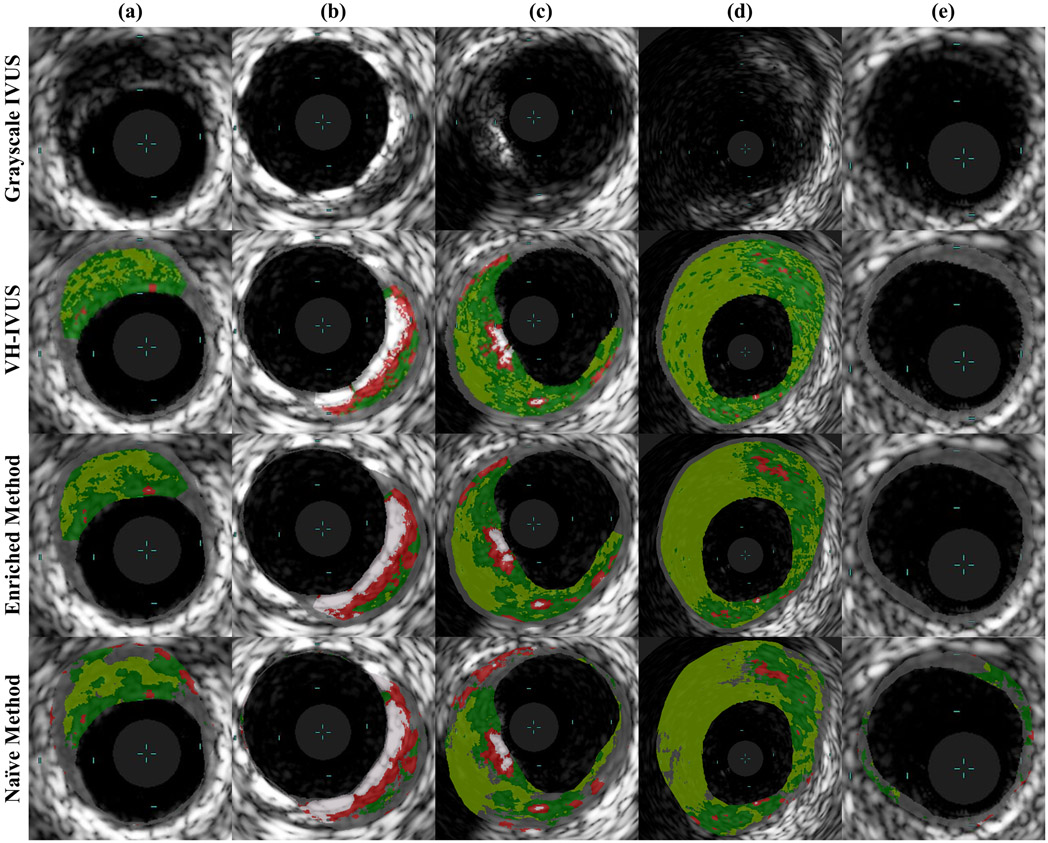
Representative examples of classified images resulting from each method are shown in Fig. 6, with detailed regions shown in Fig. 7. Both methods accurately captured major tissue morphology and features within the pathological region (Fig. 6). However, the naïve method struggled to identify non-pathological and media tissue, and occasionally generated physiologically implausible configurations (Fig. 7). Due to the spatial constrains imposed prior to CNN classification, the domain enriched method addressed non-pathological and media tissue very accurately, and was not disposed to violating physiological constraints. It captured fine features and provided sharp distinctions between various plaque types; it generated images that are very similar to gold standard VH-IVUS.
Fig. 6.
Representative classified IVUS image segmented by VH-IVUS (ground truth), naïve method, and enriched method. Both presented methods identify major pathological tissue morphology features, but the naïve method misclassifies much of the non-pathological and media tissue. The enriched method provides somewhat sharper distinctions between various plaque types and consequently captures finer features, and is most similar to VH-IVUS.
Fig. 7.
Sample classified regions of IVUS images segmented by VH-IVUS (ground truth) and the two presented methods. Both presented methods identify major pathological tissue morphology features quite well, but the enriched method demonstrates clear superiority. In these examples, the naïve method misclassifies much of the non-pathological and media tissue and proposes several variations of physiologically non-feasible morphologies. These physiological impossibilities include islands of non-pathological tissue embedded within a diseased region (A–E), exaggerated, thick segments of healthy (normally-thin) intima or media tissue (C), and calcified and lipid deposits within exceptionally thin wall segments (A, B). Light blue hash marks within each image demarcate 1 mm increments.
While the naïve method performance metrics (Table II) reflect only the five-class CNN classifier, as the classifier itself performs all segmentation operations, the overall domain enriched method metrics (Table I) depend both on (four-class) classifier performance and reliability of pathological tissue detection, which together share responsibility for the full segmentation procedure. The CNN classifiers, trained only on pixels classified by VH-IVUS, achieved generally high precision (i.e. positive predictive value) and recall (i.e. sensitivity). Table SI (in Supplemental Materials) shows the error matrices for the enriched method’s four-class CNN classifier – the model achieved an accuracy of 92.3% (cf. naïve five-class classifier accuracy of 87.8%, Table II). CNN training took several weeks (roughly 3 days per epoch for the 5-class model and somewhat less for the 4-class model). Training was halted once accuracy and loss plateaued, after no more than 50 epochs (Fig. S5); with further training, validation metrics deteriorated, indicating overfitting of the model to training data.
Error matrices of the classifiers illustrate some general and model-specific trends. Both classifiers – the five-class network supporting the naïve method and the four-class network supporting the domain enriched method – struggle to differentiate FFT from FT and, unexpectedly, DC from NC. Notably, while class confusion trends were universally observed for both models, performance was worse in all cases for the 5-class CNN except in the task of identifying calcium (DC). Furthermore, classification of the media by this model is only mediocre – pixels belonging to the M class are often misclassified as FT, FFT, or NC, and these tissues are conversely misclassified as M with moderate frequency. These findings show that imposing spatial constraints to determine non-pathological and media tissue prior to CNN classification, and excluding this class from classification, not only improved segmentation of this non-diseased tissue type, but that of the classified plaque as well. However, the enriched model was still subject to compounding uncertainties arising from pathological tissue delineation. While delineation of pathological tissue, as defined by VH-IVUS, was very accurate, the CNN of the enriched method was incapable of classifying M tissue it encountered (and typically identified it as FT; Table SI).
Execution time of the characterization method was dominated by the pixel-wise network classification of the ROI. Each pixel took 7.4 ± 0.4 milliseconds (mean ± standard deviation) to classify, though this value was found to be very sensitive to the machine on which classification was performed. Each ROI contained 37801 ± 22455 pixels, of which the enriched method determined that 26776 ± 20805 pixels were pathological and subsequently classified by the network. (The naïve method classified all pixels within the entire ROI.) Calculation of D1 and D2, and subsequent designation of the media and non-pathological tissue in a frame, took just 25.5 ± 0.9 milliseconds per frame. Because the ROI delineation method is considered interchangeable for this method, execution time of this step was not determined, but several methods report execution times significantly less than 1 second per frame [13], [14], [17], [18]. Consequently, characterization of full frames took 200 ± 150 seconds and 280 ± 170 seconds with the enriched and naïve methods, respectively. The range of execution times corresponds to the drastic variability in plaque content between frames; while segments with high plaque burden took several minutes to characterize, frames depicting cross-sections without diseased tissue (just media and/or non-pathological tissue) took just a fraction of a second for the enriched method. We note here that per-frame characterization time is reported for a scenario in which every individual pixel of the ROI is characterized, rather than a strategically selected subset, and furthermore neither software nor hardware were optimized for execution time. As such, these times should be interpreted as an upper bound.
Supplemental results, including those of a sensitivity analysis of patch size, as well as an ablation study of the enriched network’s CNN, are provided in the Supplemental Materials.
V. Discussion
The confluence of domain knowledge in vascular pathology and physiology and intravascular imaging, and advancements in machine learning, has enabled an enhanced deep learning approach to classify atherosclerosis using intravascular ultrasound grayscale images. This approach exceeds the performance of previously-reported methods for plaque segmentation in IVUS without the use of spectral signals [4], and produces maps of tissue morphology that closely resemble VH-IVUS. Of great importance, the method offers attributes that exceed those of VH-IVUS. Because no RF (spectral) data are required, the method’s applicability is not limited to ECG-gated frames, but can be used to extract plaque morphologies in any grayscale IVUS image. To acquire the same lateral resolution of plaque morphology using VH-IVUS would require extensive procedural time; the method is also not subject to the loss of temporal resolution that limits VH-IVUS [31]. Furthermore, VH-IVUS offers lower axial spatial resolution than its grayscale counterpart [7], [9], [31], suggesting that a classification method based upon the grayscale information alone could offer superior detail and information on fine features. All of these benefits are achieved without the need for specialized hardware or proprietary software.
The impact of leveraging domain knowledge to distinguish pathological from non-pathological tissue prior to CNN classification was assessed, and was found to offer substantial benefit. In particular, enforcing physiologically-imposed spatial constraints to assign the non-pathological and media tissue class not only improved classification performance for this class, but also benefited classification of the remaining pathological tissue types and decreased execution time. Application of this domain knowledge further prevented various forms of unrealistic morphologies that arose in the unconstrained naïve model. Implementing the enriched method and subjecting it to protracted training on an extensive dataset produced excellent results.
While previous methods have classified tissue in grayscale IVUS images, the method presented here surpasses performance of the current state-of-the-art. Previous work trained and validated on the same dataset implemented several varieties of classification algorithms, including support vector machines, neural networks, and random forests, with the latter achieving greatest performance. This method achieved an overall accuracy of 85.65%; sensitivity for the five classes ranged from 63.47% to 97.31%, while specificity ranged from 93.34% to 99.29% [24]. Because neural network training data can dramatically impact intravascular image segmentation performance metrics [40], direct comparison with other work is tenuous, though performance meets or exceeds all comparable methods reported in literature (Table III). Standardized datasets and methods to benchmark, analyze, and thereby fairly compare methods of intravascular tissue characterization are still needed, as has been previously established for evaluating lumen and media segmentation in IVUS by Balocco et al. [18]. To enable independent evaluation, and in anticipation of a future community standard for performance assessment, full confusion matrices have been reported here in order to allow computation of evaluation measures that are likely to be determined for such purposes.
In many ways, the benefits of applying the domain knowledge to segment the non-pathological and media tissue were foreseeable and expected. Clinical expert consensus reported by the American College of Cardiology and developed in collaboration with the European Society of Cardiology maintains that, while the trailing edge of the media (media-adventitia border) is generally well delineated in IVUS images, the leading edge is not [7]. Automated edge detection therefore only extracts lumen (lumen-intima) and media-adventitia borders, and the resulting wall area analyzed is consequently the plaque plus media area [7]. It is not surprising, then, that a CNN would have difficulty distinguishing the media from surrounding tissue within this region of a grayscale image, since the echoreflectivity profile is not conducive to distinctive transitions and the region is often not distinguishable even by trained experts. Furthermore, the spatial invariance intrinsically assumed by CNNs – generally one of their great assets in image processing – here is a liability, as the media is spatially constrained between the intima (where plaque develops) and the adventitia layers of a blood vessel. Therefore, utilizing a priori knowledge, derived previously from studies using alternative visualization modalities and mechanisms (e.g. histology [37], [38]), provided strong benefit. Furthermore, imposing geometric constraints based in physical reality made the method more robust to poor image quality and artifacts by preventing impossible class configurations. And finally, reducing the number of classes improved classification accuracy, precision, and specificity by the CNN for all but one of the remaining classes while also reducing the number of pixels to be classified, thereby decreasing execution time.
Additionally, results showed that FFT and FT were confused by both models at much higher rates than other pairs of classes. This can also be appreciated and anticipated through knowledge of the class tissue constitution. As noted before, fibro-fatty and fibrotic tissue both contain collagen fibers, but configured differently. The former contains collagen bundled in fibers [10] and collagen in the latter are loosely packed fibers embedded in lipid accumulations [11]. It is expected then that the similarities in composition would result in similar echoreflective properties that would consequently make them difficulty to distinguish from each other. Indeed, several previous methods have reported similar difficulties in distinguishing FFT or mixed tissue from FT, and some have forgone the distinction altogether and lumped several classes into larger, more easily differentiated groups [4].
Another pair of tissue classes confused with moderate frequency was NC and DC, though not in equal portions. While just over 9% of NC pixels were misclassified as DC, only around 1% of DC pixels were misclassified as NC. Further insight is offered by the ablation study performed on the CNN, which suggested that DC and NC shared features in network representation (see Supplemental Materials for details). When DC class output was inhibited, NC sensitivity increased, though the conjugate is not true. This observation prompted an investigation of activation strength for each class, which revealed that the predicted class score for calcium was, on average, 19%p higher than that for necrotic core (Table SII). Due to the strong network response invoked by calcium, mild deviation in necrotic core appearance could be enough for the response to be eclipsed. Calcified and necrotic tissue often appear in tandem, and calcified structures are associated with acoustic shadowing [7], [31]; the imbalanced misclassification phenomenon could potentially be explained by such shadowing confounding the CNN as it identifies features of necrotic core that vary in appearance depending on its spatial position relative to the calcium. Accommodating such variation may result in the overall weaker activation for individual observations of NC tissue and consequent non-reciprocated misclassification as DC.
Segmentation of the vessel’s inner and outer border, which together circumscribe the ROI, is a critical prerequisite to extract the geometric information necessary for the enrichment of the deep learning approach, and limits the accuracy of its results. This is a limitation shared with VH-IVUS; just as VH-IVUS relies upon – indeed assumes – an accurate inner and outer border to determine plaque composition within the vessel wall [11], so too does our method. This is especially true of the domain enrichment employed by our method, and media and non-pathological tissue characterization is consequently particularly sensitive to ROI delineation. Any diminished performance in the ROI delineation degrades overall vessel characterization performance and compounds the final classification error, and as such contributions of this step are included in the reported errors. Indeed, a former study of cumulative error propagation in plaque image characterization found that image formation and border detection errors contribute to and increase plaque characterization error (i.e. decrease accuracy), but that these contributions are in acceptable limits and would not affect clinical decision [41]. Furthermore, accurate automated border detection algorithms are available, and because this segmentation is an interchangeable module on which our method builds, new or specialized methods may be utilized at will in concert with the presented domain-enriched method.
Work is warranted to extend validation of this method to ground truth histology. In the present work, the methods have been both trained and validated against VH-IVUS. While VH-IVUS has itself been validation through in vitro histopathology [28], [29], it remains a step removed from the ultimate aim of classifying the tissue underlying the image. Furthermore, expert recommendations on intravascular radiofrequency data analysis maintain that media thickness cannot, in fact, be measured using either grayscale IVUS or VH-IVUS; media labels in the VH-IVUS images are themselves based on histological studies [31]. In a way, our domain enriched method emulates this approach; use of VH-IVUS for validation may therefore somewhat exaggerate the true benefit of the approach in considering the goal of tissue characterization. For example, because media thickness actually varies [31], [37], [38], a more sophisticated method of approximating media thickness (rather than assuming a fixed threshold thickness) may better reflect the underlying imaged tissue. However, in achieving the goal of replicating the utility of VH-IVUS without its associated restrictions and burdens, VH-IVUS itself presents a desirable, useful, and well-validated reference. Still, vigilance and transparency is prudent to avoid reinforcing potentially unfounded or weak assumptions that have guided development of VH-IVUS and the medical field more broadly.
Further work should also address the execution speed of the method. As currently implemented, the method cannot be applied in real time, limiting its usefulness. Immediate and drastic improvements could be achieved by exploring strategies to tactically select subsets and/or ordered progressions of pixels to be classified, rather than classifying every single pixel in the ROI sequentially by index. Updates to software, possibly including programming language, may also be accompanied by optimization of hardware.
Finally, as with any classification system, appropriateness of the model must be considered for any specific application. In particular, previous work has demonstrated that neural network training data profoundly impacts intravascular image segmentation [40]. Here, equal representation across all classes was enforced in the training dataset, and the CNN model was consequently optimized for balanced accuracy across all classes, rather than weighted by prevalence in the dataset or overall population. Therefore, other models may prove more appropriate for the detection of specific plaque types or in patient populations with plaque phenotype profiles which deviate significantly from a balanced distribution. Furthermore, IVUS images can vary significantly in texture and appearance depending on the specific imaging system (hardware and software), system settings (e.g. transducer frequency), and acquisition protocol; performance of analysis algorithms can vary commensurately [18]. Generalizability of the specific network and quantitative performance reported should not be assumed for other datasets, though general trends regarding the impact of domain enrichment are expected to hold.
VI. Conclusion
By leveraging domain knowledge and recent technological advances, a domain enriched method of classifying plaque morphology using only grayscale IVUS images has achieved higher accuracy than that of others previously reported. By first imposing geometric constrains based upon pathological studies and normal vessel morphology, segmented images have been produced that replicate VH-IVUS characterization with exceptional fidelity – without use of RF signal data. The method can therefore be applied to any grayscale IVUS data, including previously-acquired images that have not been characterized by the VH technique and images in VH-IVUS acquisitions occurring between characterized ECG-gated frames, thereby increasing the effective information acquisition speed. While care must be taken to consider and convey assumptions which may be reinforced or perpetuated through the application of domain knowledge to learning methods for medical imaging, this method offers practical, translational opportunities for immediate application-specific deployment.
Supplementary Material
Acknowledgment
The authors wish to thank the Department of Cardiology at the University of Ioannina Medical School for providing the imaging data used in this work.
This work was supported in part by the U.S. National Institutes of Health under Grant R01 49039 awarded to E. R. Edelman and in part by a NVIDIA GPU Corporation Grant awarded to L. S. Athanasiou.
Biography

Max L. Olender (M’17) was born in Detroit, MI, USA. He received the B.S.E. degree in mechanical engineering (minor in electrical engineering) in 2015 and M.S.E. degree in biomedical engineering in 2016 from the University of Michigan, Ann Arbor, MI, USA. He is currently a candidate for the Ph.D. degree in mechanical engineering at the Massachusetts Institute of Technology, Cambridge, MA, USA.
He currently works as a research assistant in the Harvard-MIT Biomedical Engineering Center, Cambridge, MA, USA, and serves as an Associate Editor for the Journal of Science Policy & Governance. He previously conducted research at the University of Michigan with the Biomechanics Research Laboratory and the Neuromuscular Lab. He has published his research in the IEEE Transactions on Medical Imaging and presented work at several international IEEE conferences with peer-reviewed conference proceedings.
Mr. Olender is a student member of the American Association for the Advancement of Science, Biomedical Engineering Society, and European Society of Biomechanics.

Lambros S. Athanasiou (M’16) was born in Ioannina, Greece in 1985. He received the engineering degree from the University of Aegean, Samos, Greece in 2009 and his Ph.D. degree from the University of Ioannina, Ioannina, Greece in 2015. In 2019, he concluded a postdoctoral research fellowship at the Massachusetts Institute of Technology, Cambridge, MA, USA and the Brigham and Women’s Hospital, Boston, MA, USA.
He has authored dozens of research articles, book chapters, and a book. His research interests include medical image processing, machine learning, biomedical engineering, and medical expert systems.
Dr. Athanasiou has served as a reviewer for several professional scientific associations and committees.

Lampros K. Michalis was born in Arta, Greece in 1960. He received the M.D. degree with distinction from the Medical School, University of Athens, Athens, Greece, in 1984, where in 1989, he was awarded his M.D. thesis with distinction.
He has been fully trained in clinical and interventional cardiology in the United Kingdom. Since 1995, he has been with the University of Ioannina Medical School, Ioannina, Greece, where he is a Professor in the Department of Cardiology. His research interests include interventional cardiology, vibrational angioplasty, intravascular imaging, coronary and peripheral artery disease, and bioengineering.

Dimitrios I. Fotiadis (M’01–SM’07–F’19) was born in Ioannina, Greece in 1961. He received the Diploma degree in chemical engineering from the National Technical University of Athens, Athens, Greece, in 1985 and the Ph.D. degree in chemical engineering from the University of Minnesota, Minneapolis, MN, USA in 1990.
He is currently a Professor of Biomedical Engineering in the Department of Materials Science and Engineering, University of Ioannina, Ioannina, Greece, where he is also the Director of the Unit of Medical Technology and Intelligent Information Systems, and is also an Affiliated Member of the Foundation for Research and Technology Hellas, Institute of Molecular Biology and Biotechnology, Department of Biomedical Research. He was a Visiting Researcher at the RWTH, Aachen, Germany, and the Massachusetts Institute of Technology, Cambridge, MA, USA. He has coordinated and participated in more than 200 R&D funded projects (in FP6, FP7, H2020, and national projects), being the coordinator and technical coordinator in several of them.
Prof. Fotiadis is an IEEE EMBS Fellow, EAMBES Fellow, Fellow of IAMBE, member of the IEEE Technical Committee of Information Technology in Healthcare, Editor-in-Chief of IEEE Journal of Biomedical and Health Informatics, Associate Editor of IEEE Reviews in Biomedical Engineering, IEEE Open Journal in Engineering in Medicine and Biology, and Computers in Biology and Medicine. His research interests include multiscale modeling of human tissues and organs, intelligent wearable/implantable devices for automated diagnosis, processing of big medical data, machine learning, sensor informatics, image informatics, and bioinformatics. He is the recipient of many scientific awards including the one by the Academy of Athens.

Elazer R. Edelman was born in New York, NY, USA. He received B.S. degrees in bioelectrical engineering and in applied biology in 1978 and the M.S. degree in electrical engineering and computer science in 1979 from the Massachusetts Institute of Technology (MIT), Cambridge, MA, USA. He received the M.D. degree from Harvard Medical School, Boston, MA, USA in 1983, and the Ph.D. degree in medical engineering and medical physics from MIT in 1984.
He is currently the Edward J. Poitras Professor in Medical Engineering and Science at MIT, Professor of Medicine at Harvard Medical School, and Senior Attending Physician in the coronary care unit at the Brigham and Women’s Hospital in Boston, MA, USA. He is also Director of MIT’s Institute for Medical Engineering and Science and Clinical Research Center, as well as the Harvard-MIT Biomedical Engineering Center.
Prof. Edelman is a fellow of the American College of Cardiology, American Heart Association, Association of University Cardiologists, American Society of Clinical Investigation, American Institute of Medical and Biological Engineering, American Academy of Arts and Sciences, National Academy of Inventors, Institute of Medicine/National Academy of Medicine, and National Academy of Engineering.
Contributor Information
Max L. Olender, Department of Mechanical Engineering and the Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA 02139 USA.
Lambros S. Athanasiou, Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA 02139 USA; Cardiovascular Division, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115 USA.
Lampros K. Michalis, Faculty of Medicine, School of Health Sciences, University of Ioannina and the 2nd Department of Cardiology, University Hospital of Ioannina, Ioannina, 45500 Greece.
Dimitris I. Fotiadis, Unit of Medical Technology and Intelligent Information Systems, Department of Materials Science and Engineering, University of Ioannina, Ioannina, 45110 Greece; Department of Biomedical Research, Institute of Molecular Biology and Biotechnology - FORTH, Ioannina, 45110 Greece..
Elazer R. Edelman, Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA 02139 USA; Cardiovascular Division, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115 USA
References
- [1].Libby P, “Inflammation in atherosclerosis,” Nature, vol. 420, no. 6917, pp. 868–874, December 2002. [DOI] [PubMed] [Google Scholar]
- [2].Bentzon JF, Otsuka F, Virmani R, and Falk E, “Mechanisms of Plaque Formation and Rupture,” Circ. Res, vol. 114, no. 12, pp. 1852–1866, June 2014. [DOI] [PubMed] [Google Scholar]
- [3].Benjamin EJ et al. , “Heart Disease and Stroke Statistics—2019 Update: A Report From the American Heart Association,” Circulation, vol. 139, no. 10, pp. e56–e528, March 2019. [DOI] [PubMed] [Google Scholar]
- [4].Athanasiou LS et al. , “Currently available methodologies for the processing of intravascular ultrasound and optical coherence tomography images,” Expert Rev. Cardiovasc. Ther, vol. 12, no. 7, pp. 885–900, July 2014. [DOI] [PubMed] [Google Scholar]
- [5].Koganti S, Kotecha T, and Rakhit RD, “Choice of Intracoronary Imaging: When to Use Intravascular Ultrasound or Optical Coherence Tomography,” Interv. Cardiol. Rev, vol. 11, no. 1, pp. 11–16, May 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bourantas CV et al. , “Hybrid intravascular imaging: current applications and prospective potential in the study of coronary atherosclerosis,” J. Am. Coll. Cardiol, vol. 61, no. 13, pp. 1369–78, April 2013. [DOI] [PubMed] [Google Scholar]
- [7].Mintz GS et al. , “American College of Cardiology clinical expert consensus document on standards for acquisition, measurement and reporting of intravascular ultrasound studies (IVUS),” J. Am. Coll. Cardiol, vol. 37, no. 5, pp. 1478–1492, April 2001. [DOI] [PubMed] [Google Scholar]
- [8].Vince DG et al. , “Automated coronary plaque characterization with intravascular ultrasound backscatter: In vivo and ex vivo validation,” J. Acoust. Soc. Am, vol. 119, no. 5, pp. 3256–3256, May 2006. [PubMed] [Google Scholar]
- [9].Garcia-Garcia HM, Costa MA, and Serruys PW, “Imaging of coronary atherosclerosis: intravascular ultrasound,” Eur. Heart J, vol. 31, no. 20, pp. 2456–2469, October 2010. [DOI] [PubMed] [Google Scholar]
- [10].Burke AP, Kolodgie FD, Farb A, Weber D, and Virmani R, “Morphological Predictors of Arterial Remodeling in Coronary Atherosclerosis,” Circulation, vol. 105, no. 3, pp. 297–303, January 2002. [DOI] [PubMed] [Google Scholar]
- [11].Konig A and Klauss V, “Virtual histology,” Heart, vol. 93, no. 8, pp. 977–982, August 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gerbaud E et al. , “Multi-laboratory inter-institute reproducibility study of IVOCT and IVUS assessments using published consensus document definitions,” Eur. Hear. J. – Cardiovasc. Imaging, vol. 17, no. 7, pp. 756–764, July 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Plissiti ME, Fotiadis DI, Michalis LK, and Bozios GE, “An Automated Method for Lumen and Media–Adventitia Border Detection in a Sequence of IVUS Frames,” IEEE Trans. Inf. Technol. Biomed, vol. 8, no. 2, pp. 131–141, June 2004. [DOI] [PubMed] [Google Scholar]
- [14].Moshfegh A, Javadzadegan A, Mohammadi M, Ravipudi L, Cheng S, and Martins R, “Development of an innovative technology to segment luminal borders of intravascular ultrasound image sequences in a fully automated manner,” Comput. Biol. Med, vol. 108, pp. 111–121, May 2019. [DOI] [PubMed] [Google Scholar]
- [15].Wang Y, Qiu C, Jiang J, and Xia S, “Detecting the Media-adventitia Border in Intravascular Ultrasound Images through a Classification-based Approach,” Ultrason. Imaging, vol. 41, no. 2, pp. 78–93, March 2019. [DOI] [PubMed] [Google Scholar]
- [16].Lo Vercio L, del Fresno M, and Larrabide I, “Lumen-intima and media-adventitia segmentation in IVUS images using supervised classifications of arterial layers and morphological structures,” Comput. Methods Programs Biomed, vol. 177, pp. 113–121, August 2019. [DOI] [PubMed] [Google Scholar]
- [17].Kim S, Jang Y, Jeon B, Hong Y, Shim H, and Chang H, “Fully Automatic Segmentation of Coronary Arteries Based on Deep Neural Network in Intravascular Ultrasound Images,” in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol. 11043 LNCS, 2018, pp. 161–168. [Google Scholar]
- [18].Balocco S et al. , “Standardized evaluation methodology and reference database for evaluating IVUS image segmentation,” Comput. Med. Imaging Graph, vol. 38, no. 2, pp. 70–90, March 2014. [DOI] [PubMed] [Google Scholar]
- [19].Zhang X, McKay CR, and Sonka M, “Tissue characterization in intravascular ultrasound images,” IEEE Trans. Med. Imaging, vol. 17, no. 6, pp. 889–899, December 1998. [DOI] [PubMed] [Google Scholar]
- [20].Brunenberg E, Pujol O, ter Haar Romeny B, and Radeva P, “Automatic IVUS Segmentation of Atherosclerotic Plaque with Stop & Go Snake,” Med. Image Comput. Comput. Assist. Interv, 2006, pp. 9–16. [DOI] [PubMed] [Google Scholar]
- [21].Athanasiou LS et al. , “A hybrid plaque characterization method using intravascular ultrasound images,” Technol. Heal. Care, vol. 21, no. 3, pp. 199–216, June 2013. [DOI] [PubMed] [Google Scholar]
- [22].Taki A et al. , “A New Approach for Improving Coronary Plaque Component Analysis Based on Intravascular Ultrasound Images,” Ultrasound Med. Biol, vol. 36, no. 8, pp. 1245–1258, August 2010. [DOI] [PubMed] [Google Scholar]
- [23].Taki A, Roodaki A, Setarehdan SK, Avansari S, Unal G, and Navab N, “An IVUS image-based approach for improvement of coronary plaque characterization,” Comput. Biol. Med, vol. 43, no. 4, pp. 268–280, May 2013. [DOI] [PubMed] [Google Scholar]
- [24].Athanasiou LS et al. , “A novel semiautomated atherosclerotic plaque characterization method using grayscale intravascular ultrasound images: Comparison with virtual histology,” IEEE Trans. Inf. Technol. Biomed, vol. 16, no. 3, pp. 391–400, May 2012. [DOI] [PubMed] [Google Scholar]
- [25].Hwang YN, Lee JH, Kim GY, Shin ES, and Kim SM, “Characterization of coronary plaque regions in intravascular ultrasound images using a hybrid ensemble classifier,” Comput. Methods Programs Biomed, vol. 153, pp. 83–92, January 2018. [DOI] [PubMed] [Google Scholar]
- [26].Kim GY, Lee JH, Hwang YN, and Kim SM, “A novel intensity-based multi-level classification approach for coronary plaque characterization in intravascular ultrasound images,” Biomed. Eng. Online, vol. 17, no. S2, November 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nair A, Kuban BD, Tuzcu EM, Schoenhagen P, Nissen SE, and Vince DG, “Coronary Plaque Classification With Intravascular Ultrasound Radiofrequency Data Analysis,” Circulation, vol. 106, no. 17, pp. 2200–2206, October 2002. [DOI] [PubMed] [Google Scholar]
- [28].Nasu K et al. , “Accuracy of In Vivo Coronary Plaque Morphology Assessment,” J. Am. Coll. Cardiol, vol. 47, no. 12, pp. 2405–2412, June 2006. [DOI] [PubMed] [Google Scholar]
- [29].Van Herck J, De Meyer G, Ennekens G, Van Herck P, Herman A, and Vrints C, “Validation of in vivo plaque characterisation by virtual histology in a rabbit model of atherosclerosis,” EuroIntervention, vol. 5, no. 1, pp. 149–156, May 2009. [DOI] [PubMed] [Google Scholar]
- [30].Nair A, Margolis MP, Kuban BD, and Vince DG, “Automated coronary plaque characterisation with intravascular ultrasound backscatter: ex vivo validation,” EuroIntervention, vol. 3, no. 1, pp. 113–20, May 2007. [PubMed] [Google Scholar]
- [31].García-García H et al. , “Tissue characterisation using intravascular radiofrequency data analysis: recommendations for acquisition, analysis, interpretation and reporting,” EuroIntervention, vol. 5, no. 2, pp. 177–189, June 2009. [DOI] [PubMed] [Google Scholar]
- [32].Athanasiou LS, Olender ML, de la Torre Hernandez JM, Ben-Assa E, and Edelman ER, “A deep learning approach to classify atherosclerosis using intracoronary optical coherence tomography,” in Medical Imaging 2019: Computer-Aided Diagnosis, 2019, vol. 10950. [Google Scholar]
- [33].Balocco S, González M, Ñanculef R, Radeva P, and Thomas G, “Calcified Plaque Detection in IVUS Sequences: Preliminary Results Using Convolutional Nets,” in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol. 11047 LNCS, 2018, pp. 34–42. [Google Scholar]
- [34].Siogkas PK, Stefanou KA, Athanasiou LS, Papafaklis MI, Michalis LK, and Fotiadis DI, “Art care: A multi-modality coronary 3D reconstruction and hemodynamic status assessment software,” Technol. Heal. Care, vol. 26, no. 1, pp. 187–193, March 2018. [DOI] [PubMed] [Google Scholar]
- [35].Ng H-F, “Automatic thresholding for defect detection,” Pattern Recognit. Lett, vol. 27, no. 14, pp. 1644–1649, October 2006. [Google Scholar]
- [36].Kass M, Witkin A, and Terzopoulos D, “Snakes: Active contour models,” Int. J. Comput. Vis, vol. 1, no. 4, pp. 321–331, January 1988. [Google Scholar]
- [37].Waller BF, “The eccentric coronary atherosclerotic plaque: Morphologic observations and clinical relevance,” Clin. Cardiol, vol. 12, no. 1, pp. 14–20, January 1989. [DOI] [PubMed] [Google Scholar]
- [38].Waller BF, Orr CM, Slack JD, Pinkerton CA, Van Tassel J, and Peters T, “Anatomy, histology, and pathology of coronary arteries: A review relevant to new interventional and imaging techniques-Part I,” Clin. Cardiol, vol. 15, no. 6, pp. 451–457, June 1992. [DOI] [PubMed] [Google Scholar]
- [39].Goodfellow I, Bengio Y, and Courville A, Deep Learning. Cambridge, MA: MIT Press, 2016. [Google Scholar]
- [40].Gowrishankar A, Athanasiou L, Olender M, and Edelman E, “Neural Network Training Data Profoundly Impacts Texture-Based Intravascular Image Segmentation,” in 2019 IEEE 19th International Conference on Bio informatics and Bioengineering (BIBE), 2019, pp. 989–993. [Google Scholar]
- [41].Athanasiou LS et al. , “Error propagation in the characterization of atheromatic plaque types based on imaging,” Comput. Methods Programs Biomed, vol. 121, no. 3, pp. 161–174, October 2015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.