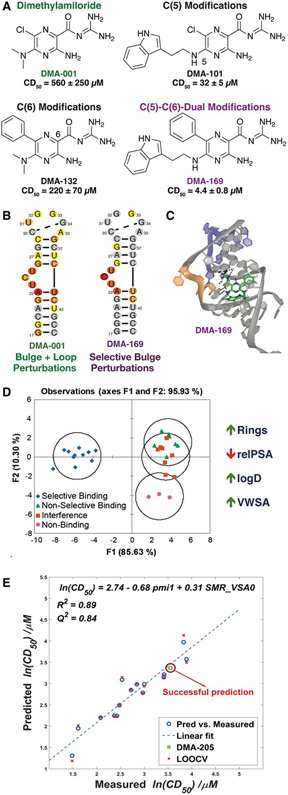
Figure 2. Dimethylamiloride (DMA) as a tunable RNA-binding scaffold.
A) Stepwise modification at the C(5) and C(6) positions of amiloride scaffold to give lead DMA-169. Competitive displacement dose (CD50) for Tat peptide assays shown below each ligand B) Heat maps of 1H-13C [HMQC] SOFAST NMR experiments with amiloride and HIV-1-TAR RNA. C) Docked pose of DMA-169 with HIV-1-TAR, which shows interactions near the trinucleotide bulge (shown in orange). D) Linear discriminate analysis based on 20 cheminformatic parameters clusters selective amiloride ligands from non-selective ligands. Sample parameters are shown to the right, with trend for selectivity indicated by the arrow. Panels A-D reproduced from Ref. 88 with permission from the Royal Society of Chemistry. E) QSAR study on ESSV ligands generated a robust model and predicted binding affinity of a new ligand (DMA-205). LOOCV = leave-one-out cross validation. Panel E reproduced from Ref. 90 with permission from the Royal Society of Chemistry.