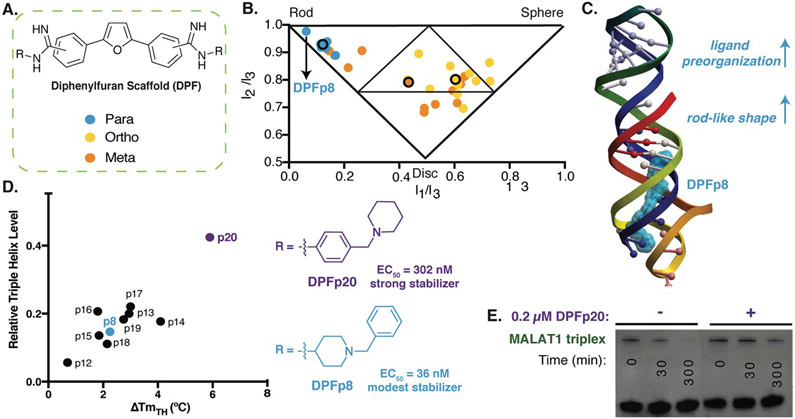
Figure 3. Diphenylfuran scaffold reveals shape-dependent RNA recognition of MALAT1 triplex.
A. Schematic of diphenylfuran (DPF) scaffold library with symmetric regio-substitution. B. Principle moments of inertia (PMI) analysis of DPF library (blue-para, orange-meta, yellow-ortho substituted) revealed a correlation between triple helix binding strengths and small molecule 3D shape, with the highest affinity triplex binder (DPFp8) as the most rod-like. Reproduced from Ref. 94 with permission from John Wiley and Sons. C. Docking model of MALAT1 triple helix structure (PDB: 4PLX, Ref. 98) with DPFp8 (blue) illustrating the importance of rod-like shape and preorganization. D) Left: Correlation of DPF-induced changes in triplex melting temperature with amount of RNA remaining in an enzyme (RNaseR) degradation assay. Right: Structures of DPFp20, the most stabilizing DPF (purple) and DPFp8, the highest affinity DPF (blue). Shown EC50 values are based on RNA titration of fluorescent DPF scaffold. E. Gel image showing stabilization of MALAT1 triplex by DPF20 to RNaseR degradation. Panels D,E reproduced from Ref. 95 with permission from Oxford University Press.