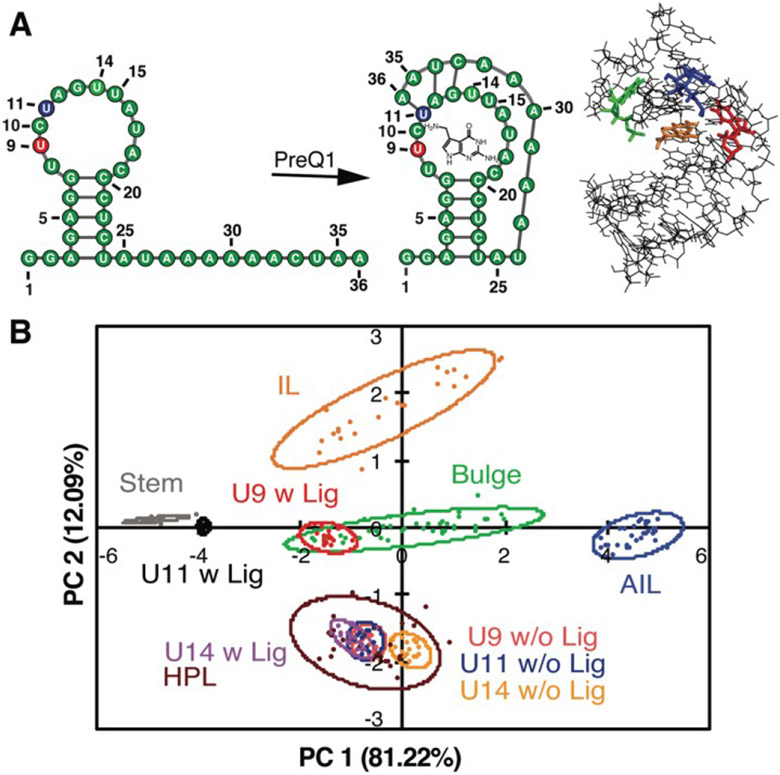
Figure 7. PRRSM classifies apo and bound secondary structures in prequeuosine-1 (PreQ1) riboswitch.
A. Left: PreQ riboswitch secondary structure in the unbound and bound state. Right: Bound structure of PreQ1 (PDB 2L1V) with highlighted sites of BFU fluorophore insertion (U9-red, U11-blue, U14-green) and PreQ1 ligand (orange). B. PCA plot of the U9, U11, and U14 modified RNA in the absence (w/o Lig) and presence (w Lig) of PreQ1 ligand. All constructs were predicted to be the correct structure in both the unbound and bound states under standard conditions (10 mM NaH2PO4, 25 mM NaCl, 4 mM MgCl2, 0.5 mM EDTA, pH 7.3 at 25 °C. Reprinted with permission from Ref. 112. Copyright 2019 American Chemical Society.