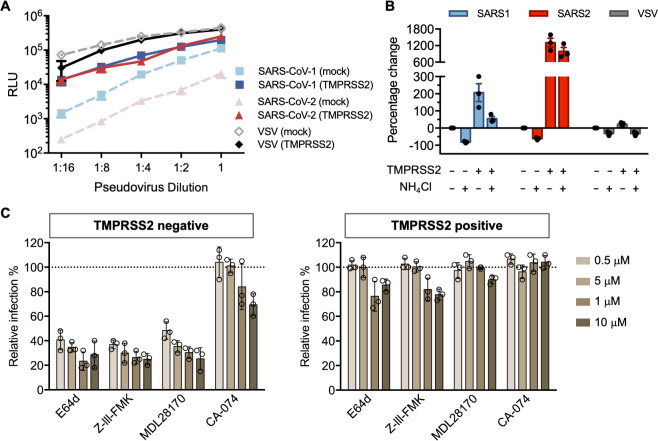
Fig 2. SARS-CoV-2 is resistant to endosomal protease inhibitors in cells over-expressing TMPRSS2.
(A) 293T-ACE2 cells were transfected with a vector control plasmid (TMPRSS2-negative), or a TMPRSS2 expression plasmid, 24 hours prior to infection. Retroviruses pseudotyped with SARS-CoV-1 (SARS1), SARS-CoV-2 (SARS2) S, and VSV G proteins were used for infection at titers yielding equivalent infection in the presence of TMPRSS2. Pseudovirus was serially diluted by 2-fold starting from stock (RLU≈250,000 for TMPRSS2-expressing cells). Cells were inoculated with diluted pseudovirus, and viral entry was determined by the luciferase activity in cell lysates within 48 hours post infection. Shown is a representative plot from three experiments. Each point indicates the mean (±SD) of duplicate samples. (B) Cells were treated by 50 mM of ammonium chloride before infection. Infection of TMPRSS2-negative cells without treatment was used for normalization. (C) A panel of cathepsin inhibitors was tested: E64d and Z-III-FMK inhibit both cathepsin B and cathepsin L; MDL28170 specifically inhibits cathepsin L, and CA-074 inhibits cathepsin B. Cells were treated with the indicated concentrations of protease inhibitors or DMSO for 2 hours, then inoculated with retrovirus harboring SARS-CoV-2 spike proteins. Luciferase activity was measured at 48 hours post inoculation. Relative infection (%) was calculated from infection of DMSO-treated cells. Each point in (B) and (C) represents the mean of triplicate samples from one experiment. Bars indicate the average of three independent experiments and error bars indicate SD.