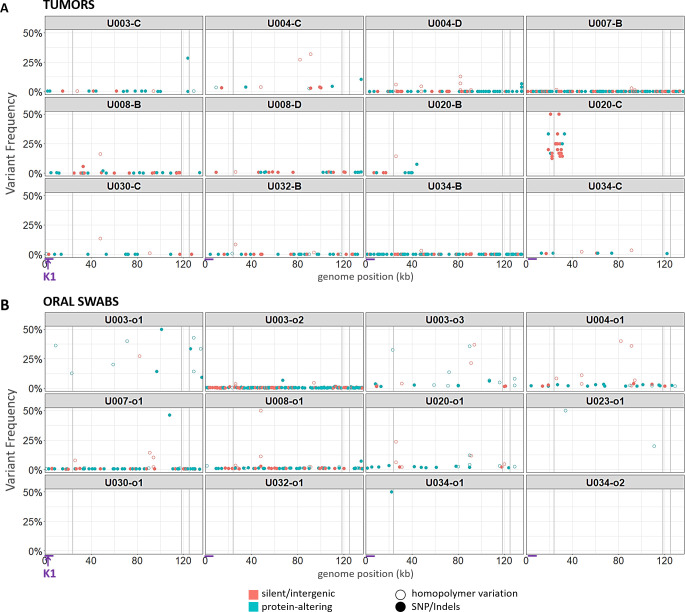
Fig 3. Point mutational diversity in KSHV genomes from tumors and oral swabs.
Bubble plots of minor sequence variants remaining after removal of PCR errors, in KSHV genomes from tumors (A) and oral swabs (B). Each bubble represents a variant base or indel, colored by whether they were predicted to be silent or protein-altering mutations. Silent mutations include synonymous and intergenic point mutations, while protein-altering mutations included non-synonymous, nonsense and frameshift mutations. Hollow circles represent mutations occurring in homopolymer runs. Bubble heights represent the frequency of the variant base among dUMI-consensus reads at that position. Vertical gray columns represent the masked repeat regions. The region containing the K1 gene is indicated with arrows at the bottom of the figure.