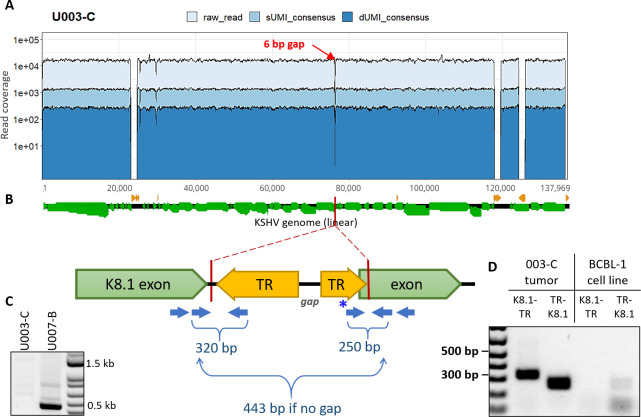
Fig 5. KSHV genomes in the U003-C tumor harbor a deletion within the K8.1 gene.
(A) Read coverage of the U003-C KSHV genome, showing a 6-bp gap (red arrow) where no read pairs were mapped. (B) Cartoon of the de novo-assembly sequences generated at either side of the gap, both ended within the K8.1 gene intron and continued into terminal repeat (TR) sequences. Green and yellow arrows show the directions of the K8.1 gene and terminal repeat sequences, respectively. Blue arrows show the position of PCR primers used to confirm breakpoint junctions, with the expected PCR product sizes. (C) PCR products generated from U003-C tumor DNA using primers flanking the gap. The 443-bp PCR product expected if the K8.1 gene intron was intact was not detected from U003-C (left column), and was detected in tumor U007-B (right column) from another person. (D) Hemi-nested PCR of U003-C tumor DNA for the K8.1-TR (left) and TR-K8.1 (right) junctions produced products of the predicted sizes. These structures were confirmed by Sanger sequencing. No K8.1-TR or TR-K8.1 junction fragment was produced from BCBL-1 DNA. The light bands at the TR-K8.1 lane under BCBL-1 were determined from Sanger sequencing to be amplicons generated from the forward primer sequence (indicated with * in panel B) overlapping with K8.1; this primer was used since the rest of the connected TR sequence assembled was GC-rich and unsuitable for primer design.