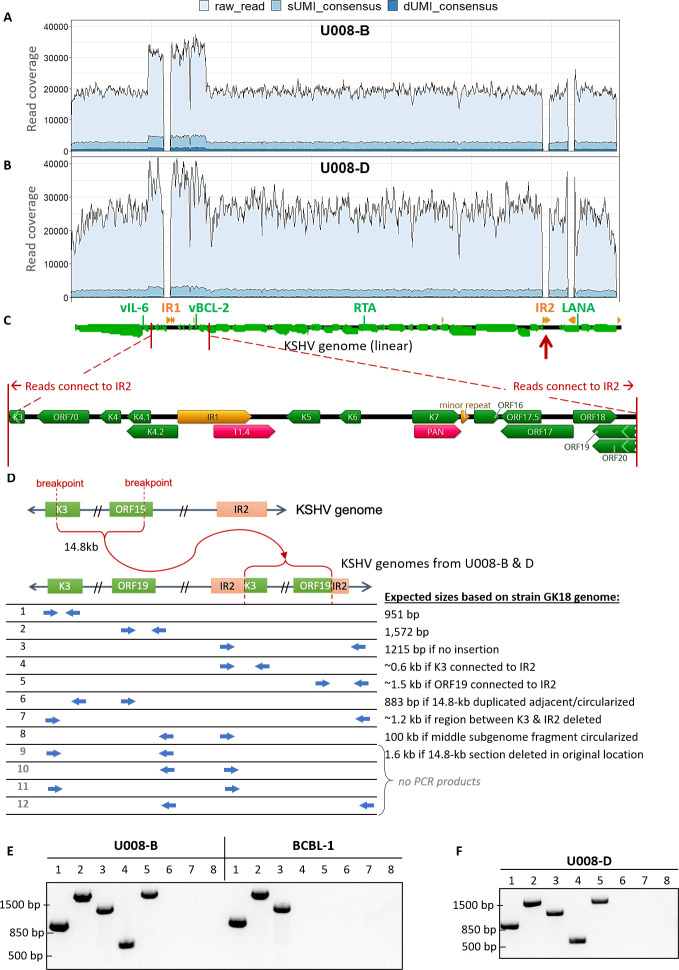
Fig 6. KSHV genomes in two tumors from participant U008 had a 14.8-kb region flanking Internal Repeat 1 (IR1) duplicated and translocated to Internal Repeat 2 (IR2).
Total, sUMI and dUMI-consensus read coverage of tumor B (A) and D (B) genomes from individual U008. (C) Annotations of the region with 1.5-2X read coverage, with genes in green, repeat regions in orange, and long non-coding RNAs in red. Many reads on the edge of this region continue into IR2 (red arrows). Annotations are from the KSHV reference isolate GK18. (D) Cartoon showing the duplication of the 14.8 kb region into IR2 and the PCR primers used to examine the genomic rearrangement in unsheared tumor DNA extracts from tumors U008-B and U008-D. PCR products produced from primer pairs numbered in D from U008-B and BCBL-1 (E) and in U008-D (F). All visible bands were excised from the agarose gel and sequenced, confirming the indicated junction sequences. Primer pairs # 9–12 produced no PCR products discernible on an agarose gel.