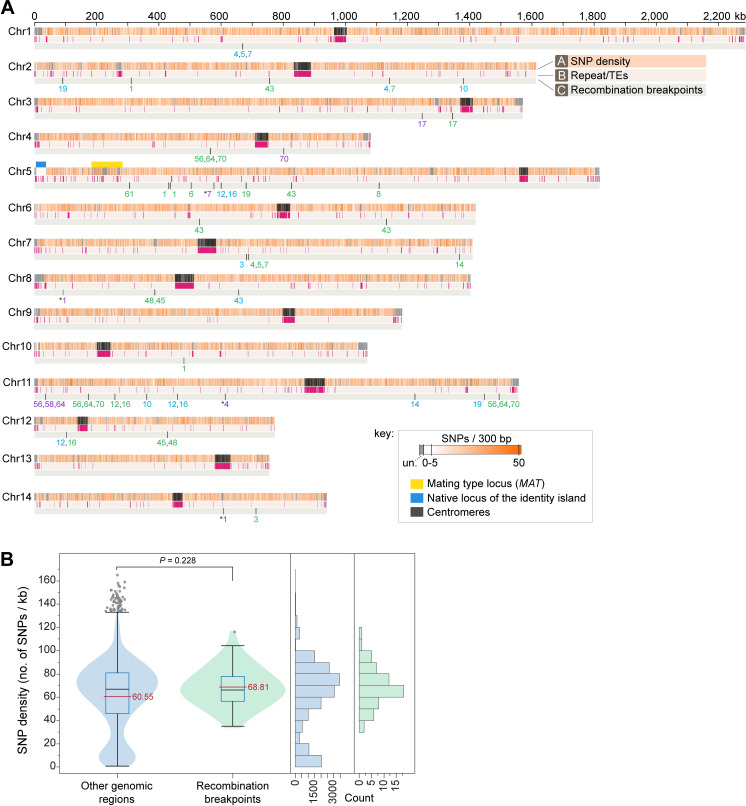
Fig 6. Recombination breakpoints identified in the hybrid progeny are not associated with repeat-rich regions nor with lower SNP density regions between the two species.
(A) Plot showing the distribution of C. deneoformans SNPs on the 14 chromosomes of C. neoformans H99 (reference), repeat content (repeats and transposable elements identified by RepeatMasker), and the location of high-confidence recombination breakpoints identified in the hybrid progeny. SNPs were calculated in 300-bp windows and plotted as a heatmap color-coded as given in the key. Regions depicted in grey represent highly divergent regions between the two reference strains and are indicated in the key as unalignable (un.). The only more closely related region (~98.5% sequence similarity) shared between the two species is indicated by a blue bar and corresponds to an ~40 kb region that resulted from a nonreciprocal transfer event (introgression) from C. neoformans to C. deneoformans [27]. The numbers below each recombination breakpoint correspond to the YX hybrid progeny strains (the YX prefix was omitted for simplicity of visualization) and are color-coded as: purple, when supported by MQ60-Cov15 and MQ60-Cov5; blue, when supported by MQ60-Cov15 and read depth; and green, when supported by MQ60-Cov15, MQ60-Cov5 and read depth (see methods for details). Some of the recombination breakpoints seem to be associated with recombination events between non-homeologous chromosomes of the two species and are marked with asterisks. (B) Violin plot, boxplots, and frequency histograms showing the SNP density within 1kb regions surrounding the high-confidence recombination breakpoints (green) compared to other genomic regions (blue). Red line, black line, blue box, and grey circles denote the mean value, median value, interquartile range, and outliers, respectively. The SNP density in the recombination breakpoint-containing regions is not statistically significantly different from the rest of the genome (Mann-Whitney test).