Abstract
Dioecy, the separation of reproductive organs on different individuals, has evolved repeatedly in different plant families. Several evolutionary paths to dioecy have been suggested, but the mechanisms behind sex determination is not well understood. The diploid dioecious Amaranthus palmeri represents a well-suited model system to study sex determination in plants. Despite the agricultural importance of the species, the genetic control and evolutionary state of dioecy in A. palmeri is currently unknown. Early cytogenetic experiments did not identify heteromorphic chromosomes. Here, we used whole-genome sequencing of male and female pools from 2 independent populations to elucidate the genetic control of dioecy in A. palmeri. Read alignment to a close monoecious relative and allele frequency comparisons between male and female pools did not reveal significant sex-linked genes. Consequently, we employed an alignment-free k-mer comparison which enabled us to identify a large number of male-specific k-mers. We assembled male-specific contigs comprising a total of almost 2 Mb sequence, proposing a XY sex-determination system in the species. We were able to identify the potential Y chromosome in the A. palmeri draft genome sequence as 90% of our male-specific sequence aligned to a single scaffold. Based on our findings, we suggest an intermediate evolutionary state of dioecy with a young Y chromosome in A. palmeri. Our findings give insight into the evolution of sex chromosomes in plants and may help to develop sustainable strategies for weed management.
Keywords: Amaranthus, dioecy, sex chromosome, invasive weed
The separation of sexes is observed in many animal and plant species, but while it is the norm in animals it is very rare in plants (Bachtrog et al. 2014). In most angiosperms, individuals have both functional sex organs and are therefore either hermaphrodite, that is, having flowers comprising the reproductive organs of both sexes, or they are monoecious, where each plant carries distinct male and female flowers. However, in a small number of plant species (≈6% of known angiosperms; Renner 2014) the female and male functions are harbored on separate individuals, that is, they are dioecious (reviewed in Charlesworth 2016). Although rare, this breeding system has evolved independently in numerous taxonomic groups with 50% of plant families having at least 1 dioecious species (Renner 2014). A potential evolutionary advantage of dioecy over cosexuality is the avoidance of inbreeding and its resulting inbreeding depression. In addition, it allows the efficient allocation of resources to specialized functions of the respective sex (Charlesworth 2016).
Sex determination in dioecious plants can be environmentally or genetically controlled (Korpelainen 1998). The genetic control of sex determination in plants ranges from few individual genes (Spigler et al. 2008; Akagi et al. 2014) to non-recombining heteromorphic sex chromosomes (Sakamoto et al. 1998; Sousa, Fuchs, and Renner 2013). Most heteromorphic systems are XY male and XX female, but female-specific sex chromosomes (ZW) also exist, for example, in Silene and Salix (Slancarova et al. 2013; Pucholt, Rönnberg-Wästljung, and Berlin 2015). Although the genetic control of dioecy has been studied in several species, the determination of sex and the evolutionary forces driving sexual dimorphism are not well understood.
Functionally, the evolution of dioecy from a cosexual ancestral state requires at least 2 mutations, 1 creating males, and 1 creating females (Westergaard 1958; Charlesworth and Charlesworth 1978). The simultaneous emergence of both mutations is highly unlikely. Hence, full dioecy has probably evolved via intermediate states, in which some individuals are female, while the others are still cosexual (Charlesworth 2016). The second mutation, which transforms the cosexual state into males by dominantly suppressing female functions, likely occurs in close linkage to the first mutation. This reduces recombination in the genomic region, as recombinant individuals would be sterile (Charlesworth 2016). In the most common case, where males are the heterozygous sex, lack of recombination between X and Y chromosomes creates a male-specific region of the Y chromosome, which can further result in gene degeneration (Ming, Bendahmane, and Renner 2011).
As the non-recombining region spreads, further degeneration is accompanied with an accumulation of transposable elements (TEs), duplications, and other mutations, leading to an increase in DNA content of the Y chromosome (Ming, Bendahmane, and Renner 2011). In plant species with heteromorphic sex chromosomes, the Y chromosome is therefore usually bigger than their X counterpart (Westergaard 1958; Sakamoto et al. 1998; Charlesworth 2013; Sousa, Fuchs, and Renner 2013). Progressing degeneration can finally lead to massive gene loss in the Y chromosome and eventually a shrinkage of the sex chromosome. This last step is common in mammals (Graves 2006) and has been observed in plants (Abraham and Mathew 1962; Segawa, Kishi, and Tatuno 1971), but appears to be less frequent (Bachtrog 2013). The above-described signature of progressive sex chromosome evolution allows the identification of the evolutionary stage of sex determination from genomic data.
The genus Amaranthus comprises over 40 species (Sauer 1957), including crops grown for grains and vegetables (Joshi et al. 2018), as well as many invasive weeds of agricultural and natural systems (e.g., Rowland, Murray, and Verhalen 1999; Bensch, Horak, and Peterson 2003). Most Amaranthus species are monoecious, while a few species exhibit a dioecious breeding system. In early classifications, dioecious Amaranthus species were grouped into a single subgenus (Acnida) based on their reproductive system (Mosyakin and Robertson 1996). Genome-wide phylogenetic work, however, indicated that these species cluster with monoecious species in distinct clades (Stetter and Schmid 2017). Hence, dioecy probably evolved multiple times independently within the Amaranthus genus.
Few weedy Amaranthus species are receiving increasing attention for their rapid evolution and spread of herbicide resistance (Kreiner et al. 2019; Molin et al. 2020). The dioecious A. palmeri L. is native to North America (Sauer 1957) and is one of the most devastating weeds in US agriculture (Webster and Nichols 2012; Riar et al. 2013). Amaranthus palmeri has evolved resistance to 8 different classes of herbicides (www.weedscience.com) and is capable of producing copious amounts of seeds (up to 600 000 per plant) (Ward, Webster, and Steckel 2013). Understanding the reproductive system and the sex determination of the species could help to develop agronomic strategies to decrease weed populations and mitigate herbicide resistances. Cytological analysis has shown that the chromosomes of male and female A. palmeri plants do not show heteromorphism (Grant 1959), suggesting an early stage of the evolution of dioecy in the species. Recent work using molecular markers suggested a genetic basis for sex determination in A. palmeri. However, due to low data quality and non-reproducible analyses, no inference of the evolutionary state of sex determination could be made (Montgomery et al. 2019).
In this study, we evaluated the evolutionary state of dioecy in A. palmeri to understand the genetic control and evolution of the separation of reproductive organs in plants. We show that dioecy in A. palmeri is controlled by a male-specific genome region suggesting an XY system. We use high depth whole genome sequencing to distinguish between a sex gene system and male-specific regions. Our high depth whole genome sequencing data allowed us to assemble a 2 Mb sex-specific region that could not be identified through allele frequency differences when aligned to a hermaphrodite relative. Alignment of our sex-specific region to a fragmented draft genome of the species identified parts of a 2 Mb scaffold as sex-specific suggesting the ongoing evolution of a non-recombining male-specific Y chromosome in A. palmeri.
Materials and Methods
Plant Material
Amaranthus palmeri seeds of 2 independent populations from California and Kansas were used. Seeds of A. palmeri from California (CA) and Kansas (KS) were cordially provided by Dr. Anil Shrestha (California State University, Fresno, California) and Dr. Dallas E. Peterson (Kansas State University, Manhattan, Kansas), respectively. We grew seeds from the 2 populations under controlled conditions in the greenhouse in Davis, Caliofonia, United States, in the summer of 2018. About 10 seeds were germinated in each plastic pot (2.37 L), filled with a soil mix (1:1 sand/peat) plus a controlled-release fertilizer (15–9–12, 150 g 75 L–1; Scotts Osmocote® PLUS). Plants were grown in a greenhouse with a temperature of 32/22ºC (day/night) and a day length of 16 h provided through supplementary lighting. Seedling were thinned randomly several times to obtain one plant per pot by their 4-leaf stage. Plants were irrigated through 4 emitters inserted into the potting medium to deliver 65 mL of water min-1 for 2 min and twice per day (7:00 am and 2:00 pm). We collected equal amounts of leaf tissue (3 disks of 5 mm diameter) of each plant once the plants were flowering and the sex could be visually determined. We pooled the leaf samples of 35 male and 32 female plants from California and 25 male and 35 female plants from Kansas for DNA extraction, creating a total of 4 pools (2 male and 2 female).
Sample Preparation and Sequencing
We extracted DNA from lyophilized and homogenized tissue using the DNeasy Plant Mini kit (Qiagen Sciences Inc, United States) and quantified DNA concentration on Qubit (Thermo Fisher Scientific, United States). Whole-genome sequencing libraries were prepared by the UC Davis Genome Center for DNA Technologies & Expression Analysis core facility using the TruSeq DNA library kit (Illumina, United States). After quality control, the samples were sequenced on an Illumina NovaSeq. We assessed the data quality using fastqc (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and estimated the sequence coverage based on the estimated genome size of 423 Mb (Stetter and Schmid 2017).
Allele Frequency Comparisons
We aligned the raw reads to the monoecious A. hypochondiracus L. reference genome v 2.1 (Lightfoot et al. 2017) using BWA-MEM version 0.7.17 (Li 2013) and removed duplicates with picard (http://broadinstitute.github.io/picard/). Then, we called SNP using GATK version 4.0.11 (McKenna et al. 2010) with the filter expression QD < 2.0 || FS > 60.0 || MQ < 40.0 || MQRankSum < -12.5 || ReadPosRankSum < -8.0 and further filtered to keep only biallelic SNPs with a maximum of 30% missing values at a site using VCFtools 0.1.16 (Danecek et al. 2011). We created a variant table using VariantsToTable function of GATK to test for allele frequency differences between female and male pools using the QTLseqR package (Mansfeld and Grumet 2018).
Reference Free k-Mer Analysis
For the reference-free comparison between male and female individuals we counted unique k-mers in the male and female pool from California. We first trimmed the reads based on their quality using trimmomatic (Bolger, Lohse, and Usadel 2014) with the parameters: SLIDINGWINDOW:4:15 MINLEN:35 LEADING:5 TRAILING:5, before counting the occurrences 35-mers in the quality trimmed sequencing reads from female and male pools separately, keeping only k-mers with a frequency between 15 and 2000 using Jellyfish v 2.3.0 (Marçais and Kingsford 2011). We combined the male and female counts to identify sex specific k-mers using custom scripts (10.6084/m9.figshare.12326306). We further, counted sex specific k-mers identified in the Californian population in the pools of the Kansas population using jellyfish query. To further generate the most robust set of k-mers, we filtered sex specific k-mers requiring a k-mer to have a count ≥ 50 but ≤ 500 within the focal Kansas (e.g., female pool for female-specific) and ≤ 20 in the Kansas opposite pool (e.g., male pool for female-specific)
Male-Specific Read Recovery
We selected male-specific k-mers that had a count of minimum 50 in the male pool and 0 (represents ≤ 15 or ≥ 2000, due to previous filtering) in the female pool. For all 4 sequenced pools, we extracted full size reads containing the k-mer sequences and their respective read pair from the trimmed read data using a custom bash script (https://doi.org/10.6084/m9.figshare.12326306).
Extraction of Male k-Mer Containing Reads From A. tuberculatus
We extracted the filtered set of male-specific k-mers from published A. tuberculatus samples with known sex expression (Kreiner et al. 2019), proceeding as described above in “male-specific read recovery.” To normalize the numbers we devided the number of extracted reads by the total number of sequenced reads.
Estimation of the Size of Male-Specific Regions
To estimate the size of the male-specific region, we assembled the recovered male-specific reads from the male California pool with platanus-allee v 2.0.2 (Kajitani et al. 2019) using default parameters. We counted the total length of assembled base pairs as sex-specific region.
Identification of Y Chromosome in A. palmeri Draft Genome
We aligned assembled male-specific scaffolds to the A. palmeri draft genome v 1.1 (Montgomery et al. 2020) using minimap2 (Li 2018) with -ax asm5 and extracted all covered sites using samtools 1.9 depth (Li et al. 2009). In addition, we mapped all extracted male and female-specific reads directly to the A. palmeri draft genome using BWA-MEM (Li 2013) and samtools to extract read depths for the male and female pool. We evaluated the sequencing depth of base pairs covered at least 50x to remove noise, as average coverage sequenced was over 300.
Results
High Coverage WGS of Male and Female Pools
We applied high depth whole genome sequencing to 4 pools of A. palmeri from 2 independent wild populations, separated by sex (i.e., 2 populations × 2 sexes). The pools consisted of combined leaf samples of 35 male and 32 female individuals from a single population collected from California, as well as 25 male and 35 female individuals from another population collected from Kansas. The per pool sequence coverage ranged from 228X to 331X (322,106,487 to 467,707,106 reads per pool), corresponding to a mean coverage of 7.5X to 9.5X per sample (Supplementary Table S1).
No Allele Frequency Differences Between Male and Female Pools
Given the lack of cytological evidence for heteromorphic sex chromosomes in A. palmeri (Grant 1959), we tested whether sex determination in A. palmeri could be controlled by a specific small and still recombining region, located on an autosome. To assess this possibility, we called biallelic SNPs and compared allele frequencies between the male and female pools of the 2 populations and tested for frequency differences. We aligned raw reads to the high-quality chromosome level assembly of the hermaphrodite A. hypochondiracus (Lightfoot et al. 2017). Overall, reads mapped to the reference genome with 90.54% to 92.55% uniquely mapped reads, indicating a high similarity between the genomes of the 2 species (Supplementary Table S1). Yet, a large number of read pairs showed non-proper paring, indicating structural differences between the genomes.
Sex determining loci in the genome are expected to lead to significant allele frequency differences between male and female pools. We used the G’ method implemented in QTLseqR to scan the first 16 Scaffolds of the reference genome, representing the 16 chromosomes of A. hypochondriacus, for significant differences in allele frequencies. Using over 12 million biallelic sites (3.2 million after filtering), distributed across the genome, we found no significant allele frequency differences between male and female pools in either of the 2 populations (Figure 1 and Supplementary Figure S1). Allele frequency differences between sexes were very low across the whole aligned genome sequence.
Figure 1.
Differences in allele frequencies between male and female pools of the California population along the genome, relative to the A. hypochondiracus reference. Positive values would represent an increased frequency of the male allele, while negative an increased frequency of the female frequency. Red and blue lines represent 95% and 99% confidence intervals for frequency outliers.
Large Number of Male-Specific k-Mers
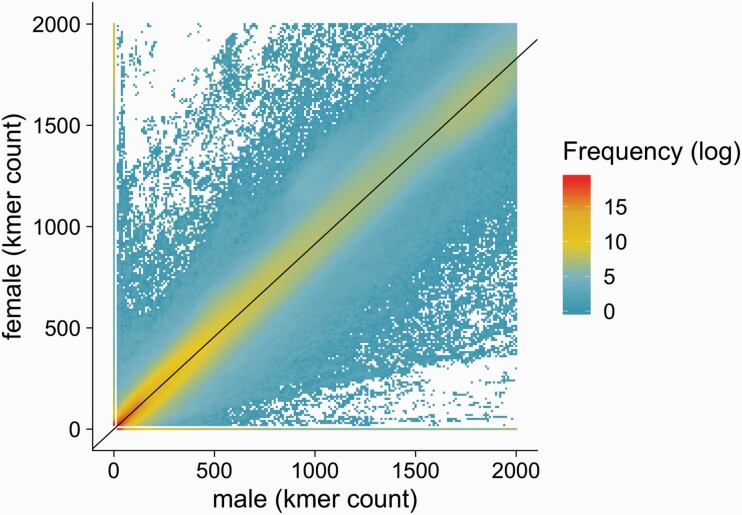
Comparing allele frequency differences requires the successful alignment of reads to the reference genome. However, sequencing reads from diverged sex-specific sequences are unlikely to align to the hermaphrodite reference genome as they accumulate large numbers of mutations (Ming, Bendahmane, and Renner 2011; Charlesworth 2013). To test whether a larger, non-recombining, region could be responsible for sex determination in A. palmeri, we compared alignment-free k-mer counts of male and female pools. We recovered all 35-mers (in the following referred to as k-mers) from the sequencing reads and counted the occurrences of these k-mers for each pool. Low frequency (≤15) and high frequency (≥2000) were discarded from the analysis as they potentially represent sequencing errors and repeat regions. We identified over 1.3 billion (1,329,452,869) male and over 1.2 billion (1,235,093,537) female k-mers that passed out filters and were used for further analysis. We compared k-mer counts of the male and female pools from the California population to find k-mers that strongly deviated from the expected equal frequency in male and female pools (Figure 2). As expected, most k-mers were present in comparable numbers within males and females (Figure 2). We found 1,634,859 k-mers that were not present in females (15≥ n ≥ 2000), but had a count between 50 and 500 in the male pool. We used the Kansas population to obtain the most robust set of specific k-mers. That is, we counted and identified sex specific k-mers for the California population in the Kansas population. We further filtered male (or female) specific k-mers for a minimum count of 50 and a maximum of 500 in the Kansas pool (or female). To be considered sex-specific, we further required a count below 20 k-mers in the female (or male) Kansas pool. After these stringent filters, we found 158,693 robust male-specific k-mers, which represent 9.7 % of all male-specific k-mers (Supplementary Table S1). We also found a low number of female-specific k-mers (269,157) in the Californian population. After correction with the Kansas population, we find 2,572 female-specific k-mers (1.0%, Supplementary Table S1). These include few k-mers, that are present over 2000 times in the male pools and therefore not counted in males.
Figure 2.
Comparison of the abundances of k-mers from the male and female sequence read pools of the Californian A. palmeri population. The color scale indicates the number distinct k-mers represented by 1 dot. The black line displays the null expectation for k-mer frequency in males and females, corrected for the number of individuals present in the male (n = 35) and female (n = 32) pool.
Extracting Male-Specific A. palmeri k-Mers From A. tuberculatus
To evaluate the age of male-specific regions and the parallelism between dioecious Amaranthus species, we extracted reads with our set of male-specific k-mers from whole-genome sequencing data of A. tuberculatus individuals (Kreiner et al. 2019), another dioecious Amaranthus species, and compared the number of extracted reads (Supplementary Figure S2). We found no significant difference between male and female individuals, suggesting that different genomic regions control dioecy in A. tuberculatus.
Identification of Y Chromosome in A. palmeri Draft Sequence
We extracted all reads with identified male-specific k-mers from the original read data to assemble the male-specific genome region. Only k-mers that were classified as male-specific in both populations were used for the extraction of reads. The assembly process resulted in 5774 contigs, which together consisted of 2,002,103 bp. The shortest contig was 151 bp long and the longest scaffold was 11,654 bp long (Supplementary Figure S3). After filtering out contigs with length below 152 (maximum length of a single read), we found 3893 contigs comprising a total length of 1,748,724 bp. Based on the estimated genome size of 423 Mb (Stetter and Schmid 2017), the identified male-specific region represents roughly 0.41% of the A. palmeri genome.
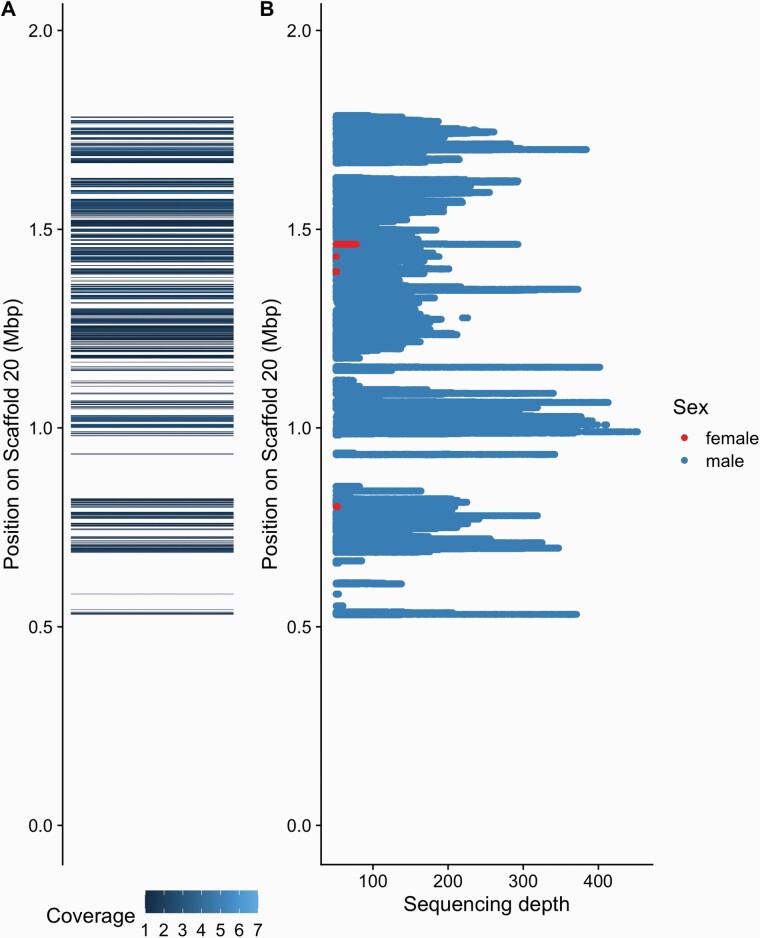
The availability of a recent draft sequence of a male A. palmeri individual (Montgomery et al. 2020) allowed us place our male-specific region and extract a potential Y chromosome. We first aligned the assembled male-specific contigs to the draft sequence. Of the 5774 contigs 55% properly aligned to the draft genome and covered a total of 617,820 bp on the draft sequence. Of all mapped male-specific base pairs that aligned, 89% mapped to a single scaffold, scaffold 20. The male-specific sequence covered 26% (510,926 bp) of bp on that scaffold (Figure 3A) and localized to a region between 0.5 Mbp and 1.9 Mbp. The first quarter of that scaffold was not covered by male-specific contigs. Other sequences mapped across 41 of the 303 scaffolds and covered less than 10% of other scaffolds.
Figure 3.
Alignment of male-specific sequences to Scaffold 20 of the draft genome sequence of a male A. palmeri. (A) Alignment of assembled male-specific regions to draft genome sequence. (B) Mapping depth of male (blue) and female (red) specific reads to the draft sequence. Showing sites with depth ≥ 50X depth.
In addition to the assembled contigs, we directly mapped sex-specific reads extracted after k-mer filtering. Again, the great majority (91%) of male-specific reads, mapped to scaffold 20, covering 22% (428,904) of this scaffold (Figure 3 and Supplementary Table S2). The reads covered a similar area of scaffold 20 as the mapping of the assembled contigs (Figure 3B). Female-specific reads only covered 501 bp on the draft sequence and 63% mapped to scaffold 20 and covered 308 bp (0.2%) and 24 % to scaffold 4 covering 125 bp of the 22 Mb scaffold.
Discussion
Male-Specific Genome Region Determines Sex
The emergence of sex chromosomes is an evolutionary process that starts with the occurrence of single mutations leading to unisexual individuals and ultimately giving rise to highly heteromorphic chromosomes. Our results show that the genome of A. palmeri harbors a male-specific region, genetically controlling the sex determination. The allocation of over 90% of male-specific reads to one part of a single scaffold (scaffold 20) suggests that the male-specific region does not recombine anymore with the analogous region on the X chromosome and has diverged sufficiently to not align to the hermaphrodite relative. The lack of alignment to the A. hypochondriacus reference sequence consequently did not allow to detect allele frequency differences between pools. Although the male sequence is strongly diverged leading to recombination suppression between male and female chromosomes in the male-specific region, an accumulation of large amounts of repetitive sequences seems to not have occurred, as it would have led to an increase in chromosome size that could be cytologically identified. Such a cytological difference has not been observed in A. palmeri (Grant 1959).
To further characterize the evolutionary state of sex determination in the species, we estimated the size of the male-specific region. Our assembly of the region suggests approximately 2 Mb. This size likely underestimates the true size of the male-specific region. Our filtering removes highly redundant k-mers, as created by TEs (Michael 2014). Repetitive sequence is also present in other parts of the genome and would therefore not be classified as males specific. The high number of contigs potentially results from gaps due to non-specificity of the sequence, however, the stringent filtering across 2 independent populations revealed high confidence sequence specific to male individuals. The alignment of our assembly to the recent draft genome sequence even allowed us to identify a single scaffold as the potential Y chromosome. The aligned male-specific regions span approximately 1.5 Mb, although not all sequence is in the region is covered (Figure 3). The gaps between male-specific regions are likely regions that are not strongly diverged from the X chromosome and therefore not male-specific.
In contrast to the male-specific k-mers, almost all female-specific k-mers found in the California sample were removed after filtering for overlaps with the Kansas population and female-specifc reads only covered very small regions of the draft sequence (Figure 3 and Supplementary Table S3). This confirms an XY system for A. palmeri as found by male-specific k-mers. However, a very low number of apparently female-specific k-mers persisted after filtering and map to a small number of scaffolds in the A. palmeri genome (Supplementary Table S3). Their low number and the fact that only a total of 501 bp are covered by such regions (mostly within the Y region) suggest them to be technical artifacts. Otherwise, they might be involved in female reproductive functions and could be further analyzed in molecular studies.
Evolution of Dioecy in A. palmeri
The different stages of sex chromosome evolution may give us insight into the question as to when dioecy has evolved in a species. For example, in the wild strawberry, Fragaria virginiana male individuals coexist with females and hermaphrodites. Full dioecy is not yet evolved and the sex-determining regions recombine to create unisexuals, hermaphrodites, and even neuters (Spigler et al. 2008). This early stage of sex chromosome evolution indicates that the separation of sexes is rather young, whereas, the sex chromosomes in mammals and birds are very old (Cortez et al. 2014; Zhou et al. 2014). The Y (mammals) and W (birds) chromosomes of the heterogametic individuals of these taxa have undergone gene loss processes and finally became smaller than their X and Z chromosomes, respectively. Most heteromorphic sex chromosomes in flowering plants are increased in size as they accumulated TEs and other repetitive sequences. It is assumed, that this stage precedes gene loss (Ming, Bendahmane, and Renner 2011). Our results show that the male-specific region in A. palmeri is strongly diverged from the X region and spans around two-thirds of scaffold 20, whilst cytological studies have not found a dimorphism between X and Y (Grant 1959). Therefore, we conclude that A. palmeri is at an early or intermediate stage of sex chromosome evolution.
The presence of cytologically homomorphic sex chromosomes that are nonetheless heteromorphic on the molecular scale has been shown in several other plant species (Liu et al. 2004; Khattak, Torp, and Andersen 2006; Telgmann-Rauber et al. 2007), such as Spinacia oleracea, belonging to the same family as A. palmeri, that is, Amaranthaceae (Khattak, Torp, and Andersen 2006).
Within the genus Amaranthus, A. palmeri and its dioecious sister species A. arenicola are placed in a monophyletic group (Stetter and Schmid 2017), in which all other species show monoecy (Wulff 1988; Costea, Sanders, and Waines 2001; Asha, Rekha, and Sadiq 2016). This suggests that dioecy has evolved independently within this group and is unlikely to be linked to the dioecious systems present in other Amaranthus species. To confirm this, we compared A. palmeri male-specific k-mers to A. tuberculatus male and female individuals and found no association with sex in this species (Supplementary Figure S2), suggesting an independent evolution of dioecy in A. palmeri. This result agrees with the phylogentic distance between A. palmeri and A. tuberculatus, which both cluster with several hermaphrodite species in genome-wide phylogenetic analyses (Stetter and Schmid 2017), but is in contrast to the earlier taxonomic classification of the subgenus acnida (Mosyakin and Robertson 1996). Based on the phylogenetic relationship the sexual dimorphism in A. tuberculatus (and A. australis and A. floridanus) is older than dioecy in A. palmeri.
Our work constitutes the first step in understanding the underlying processes of sex determination in A. palmeri, but the exact mechanisms and a potential interaction with environmental factors that has recently been suggested (Mesgaran, Matzrafi, and Ohadi 2019) remain to be discovered. The male-specific k-mers and male-specific scaffolds of the draft genome identified here, allow to diagnose the sex of individuals in early developmental stages. This permits future studies of environmental dependent sex alteration, as it will allow to distinguish between “genetic” sex and expressed sex of an individual plant. Furthermore, the assembled 2 Mb male-specific region in combination with the identified candidate Y chromosome can be used to fully assemble the A. palmeri Y chromosome. The different age and independence of dioecy evolution within the Amaranthus genus, make them a compelling system to understand the emergence of sexual dimorphism in plants. Further understanding of the sex determination in this invasive weed, can help develop novel ecological methods (e.g., shifting sex) to disrupt seed production in A. palmeri and hence reduce the impact of this invasive weed in cropping system.
Supplementary Material
Acknowledgments
We thank Julia Kreiner for sharing records of sex expression of sequenced A. tuberculatus samples. We thank Sara Ohadi for discussion and support with experimental plant care.
Funding
M.B.M. acknowledges the support from UC Davis New Research Initiatives and Interdisciplinary Research Grant and United States Department of Agriculture, National Institute of Food and Agriculture (USDA-NIFA) Hatch Funding (Project No. CA-D-PLS-2511-H). We acknowledge the support of the Deutsche Forschungsgemeinschaft under Germany’s Excellence Strategy—EXC-2048/1—Project ID 390686111 to M.G.S.
Conflict of Interest
The authors have no conflict of interest.
Author Contribution
M.B.M., M.M., and M.G.S. designed and planned the study, M.M. and M.B.M. provided plant material, M.M. and M.G.S. sampled and pooled leaf samples, A.L. performed molecular work, C.J.N. and M.G.S. analyzed the data. M.G.S. supervised the study, and M.T. and M.G.S. wrote the manuscript. All authors read and approved the manuscript.
Data Availability
Sequencing data is available from the European Nucleotide Archive (ENA) under the project numbers PRJEB38372. Scripts used for the analysis are available through figshare https://doi.org/10.6084/m9.figshare.12326306.
References
- Abraham A, Mathew PM. 1962. Cytological studies in the cycads: sex chromosomes in cycas. Ann Bot (Lond). 26:261–266. [Google Scholar]
- Akagi T, Henry IM, Tao R, Comai L. 2014. A y-chromosome–encoded small RNA acts as a sex determinant in persimmons. Science 346:646–650. [DOI] [PubMed] [Google Scholar]
- Asha S, Rekha R, Mohamed Sadiq A. 2016. Amaranthus spinosus-a review. Bull. Environ. Pharmacol. Life Sci. 5:102–107. [Google Scholar]
- Bachtrog D. 2013. Y-Chromosome evolution: emerging insights into processes of y-chromosome degeneration. Nat Rev Genet. 14:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, Ashman T-L, Hahn MW, Kitano J, Mayrose I, Ming R, et al. 2014. Sex determination: why so many ways of doing it? PLoS Biol. 12:e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensch CN, Horak MJ, Peterson D. 2003. Interference of redroot pigweed (Amaranthus retroflexus), palmer amaranth (A. palmeri), and common waterhemp (A. rudis) in soybean. Weed Sci. 51:37–43. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. 1978. A model for the evolution of dioecy and gynodioecy. Am Naturalist. 112:975–997. [Google Scholar]
- Charlesworth D. 2016. Plant sex chromosomes. Annu Rev Plant Biol. 67:397–420. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. 2013. Plant sex chromosome evolution. J Exp Bot. 64:405–420. [DOI] [PubMed] [Google Scholar]
- Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, Gruetzner F, Kaessmann H. 2014. Origins and functional evolution of Y chromosomes across mammals. Nature 508:488–493. [DOI] [PubMed] [Google Scholar]
- Costea M, Sanders A, Waines G. 2001. Preliminary results toward a revision of the Amaranthus hybridus species complex (Amaranthaceae). Sida, Contributions to Botany 19:931–974. [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant WF. 1959. Cytogenetic studies in Amaranthus: I. cytological aspects of sex determination in dioecious species. Can J Bot. 37:413–417. [Google Scholar]
- Graves JAM. 2006. Sex chromosome specialization and degeneration in mammals. Cell 124:901–914. [DOI] [PubMed] [Google Scholar]
- Joshi DC, Sood S, Hosahatti R, Kant L, Pattanayak A, Kumar A, Yadav D, and Stetter MG. 2018. From zero to hero: the past, present and future of grain amaranth breeding. Theor Appl Genet. 131:1807–1823. [DOI] [PubMed] [Google Scholar]
- Kajitani R, Yoshimura D, Okuno M, Minakuchi Y, Kagoshima H, Fujiyama A, Kubokawa K, Kohara Y, Toyoda A, Itoh T. 2019. Platanus-Allee is a de novo haplotype assembler enabling a comprehensive access to divergent heterozygous regions. Nat Commun. 10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattak JZK, Torp AM, Andersen SB.. 2006. A genetic linkage map of Spinacia oleracea and localization of a sex determination locus. Euphytica 148:311–318. [Google Scholar]
- Korpelainen H. 1998. Labile sex expression in plants. Biol Rev. 73:157–180. [Google Scholar]
- Kreiner JM, Giacomini DA, Bemm F, Waithaka B, Regalado J, Lanz C, Hildebrandt J, et al. 2019. Multiple modes of convergent adaptation in the spread of glyphosate-resistant Amaranthus tuberculatus. Proc Natl Acad Sci USA. 116:21076–21084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot DJ, Jarvis DE, Ramaraj T, Lee R, Jellen EN, Maughan PJ. 2017. Single-molecule sequencing and Hi-C-based proximity-guided assembly of amaranth (Amaranthus hypochondriacus) chromosomes provide insights into genome evolution. BMC Biol. 15:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv:1303:3997. [Google Scholar]
- Li H. 2018. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34 (18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Moore PH, Ma H, Ackerman CM, Ragiba M, Yu Q, Pearl HM, Kim MS, Charlton JW, Stiles JI, et al. 2004. A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature 427:348–352. [DOI] [PubMed] [Google Scholar]
- Mansfeld BN, Grumet R. 2018. QTLseqr: An R package for bulk segregant analysis with next-generation sequencing. The Plant Genome 11:1–5. [DOI] [PubMed] [Google Scholar]
- Marçais G, Kingsford C. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-Mers. Bioinformatics 27:764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA Sequencing Data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesgaran MB, Matzrafi M, Ohadi S. 2019. Sex lability and dimorphism in diecious palmer amaranth (Amaranthus Palmeri). bioRxiv 769935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP. 2014. Plant genome size variation: bloating and purging DNA. Briefings in Functional Genomics 13:308–317. [DOI] [PubMed] [Google Scholar]
- Ming R, Abdelhafid B, Susanne SR. 2011. Sex chromosomes in land plants. Annu Rev Plant Biol. 62:485–514. [DOI] [PubMed] [Google Scholar]
- Molin WT, Allison Y, Blenner MA, Saski CA. 2020. The eccDNA Replicon: a heritable, extra-nuclear vehicle that enables gene amplification and glyphosate resistance in Amaranthus palmeri. The Plant Cell. 32:2132–2140. doi: 10.1105/tpc.20.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery JS, Giacomini D, Waithaka B, Lanz C, Murphy BP, Campe R, Lerchl J, Landes A, Gatzmann F, Janssen A, et al. 2020. Draft genomes of Amaranthus tuberculatus, Amaranthus hybridus and Amaranthus palmeri. Genome Biology and Evolution. 12:1988–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery JS, Sadeque A, Giacomini DA, Brown PJ, Tranel PJ. 2019. Sex-specific markers for waterhemp (Amaranthus tuberculatus) and palmer amaranth (Amaranthus palmeri). Weed Sci. 67:412–418. [Google Scholar]
- Mosyakin SL, Robertson KR. 1996. New infrageneric taxa and combinations in Amaranthus (Amaranthaceae). Ann Bot Fenn. 33:275–281. [Google Scholar]
- Pucholt P, Rönnberg-Wästljung AC, Berlin S. 2015. Single locus sex determination and female heterogamety in the basket willow (Salix viminalis L.). Heredity (Edinb). 114:575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner SS. 2014. The relative and absolute frequencies of angiosperm sexual systems: dioecy, monoecy, gynodioecy, and an updated online database. Am J Bot. 101:1588–1596. [DOI] [PubMed] [Google Scholar]
- Riar DS, Norsworthy JK, Steckel LE, Stephenson DO, Eubank TW, Scott RC. 2013. Assessment of weed management practices and problem weeds in the midsouth united states—soybean: a consultant’s perspective. Weed Technol. 27:612–622. [Google Scholar]
- Rowland MW, Murray DS, Verhalen LM. 1999. Full-season palmer amaranth (Amaranthus palmeri) interference with cotton (Gossypium hirsutum). Weed Sci. 47:305–309. [Google Scholar]
- Sakamoto K, Akiyama Y, Fukui K, Kamada H, Satoh S. 1998. Characterization; genome sizes and morphology of sex chromosomes in hemp (Cannabis Sativa L.). Cytologia 63:459–464. [Google Scholar]
- Sauer J. 1957. Recent migration and evolution of the dioecious amaranths. Evolution 11:11–31. [Google Scholar]
- Segawa M, Kishi S, Tatuno S. 1971. Sex chromosomes of cycas revoluta. The Japanese Journal of Genetics 46:33–39. [Google Scholar]
- Slancarova V, Zdanska J, Janousek B, Talianova M, Zschach C, Zluvova J, Siroky J, Kovacova V, Blavet H, Danihelka J, et al. 2013. Evolution of sex determination systems with heterogametic males and females in silene. Evolution 67:3669–3677. [DOI] [PubMed] [Google Scholar]
- Sousa A, Fuchs J, Renner SS. 2013. Molecular cytogenetics (FISH, GISH) of Coccinia grandis: a ca. 3 myr-old species of cucurbitaceae with the largest Y/autosome divergence in flowering plants. Cytogenet Genome Res. 139:107–118. [DOI] [PubMed] [Google Scholar]
- Spigler RB, Lewers KS, Main DS, Ashman TL. 2008. Genetic mapping of sex determination in a wild strawberry, Fragaria virginiana, reveals earliest form of sex chromosome. Heredity (Edinb). 101:507–517. [DOI] [PubMed] [Google Scholar]
- Stetter MG, Schmid KJ. 2017. Analysis of phylogenetic relationships and genome size evolution of the Amaranthus genus using GBS indicates the ancestors of an ancient crop. Mol Phylogenet Evol. 109:80–92. [DOI] [PubMed] [Google Scholar]
- Telgmann-Rauber A, Jamsari A, Kinney MS, Pires JC, Jung C. 2007. Genetic and physical maps around the sex-determining m-locus of the dioecious plant asparagus. Mol Genet Genomics. 278:221–234. [DOI] [PubMed] [Google Scholar]
- Ward SM, Webster TM, Steckel LE. 2013. Palmer amaranth (Amaranthus palmeri): a review. Weed Technol. 27:12–27. [Google Scholar]
- Webster TM, Nichols RL. 2012. Changes in the prevalence of weed species in the major agronomic crops of the Southern United States: 1994/1995 to 2008/2009. Weed Sci. 60:145–57. [Google Scholar]
- Westergaard M. 1958. The mechanism of sex determination in dioecious flowering plants. In Advances in genetics. Vol. 9. New York: Elsevier. p. 217–281. [DOI] [PubMed] [Google Scholar]
- Wulff RD. 1988. Intraspecific variation in germination requirements and growth in Amaranthus dubius. Am J Bot. 75:1307–1312. [Google Scholar]
- Zhou Q, Zhang J, Bachtrog D, An N, Huang Q, Jarvis ED, Gilbert MT, Zhang G. 2014. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science. 346:1246338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data is available from the European Nucleotide Archive (ENA) under the project numbers PRJEB38372. Scripts used for the analysis are available through figshare https://doi.org/10.6084/m9.figshare.12326306.